Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions
- PMID: 22711839
- PMCID: PMC3396481
- DOI: 10.1073/pnas.1206999109
Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions
Abstract
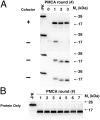
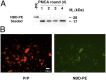
Prions containing misfolded prion protein (PrP(Sc)) can be formed with cofactor molecules using the technique of serial protein misfolding cyclic amplification. However, it remains unknown whether cofactors materially participate in maintaining prion conformation and infectious properties. Here we show that withdrawal of cofactor molecules during serial propagation of purified recombinant prions caused adaptation of PrP(Sc) structure accompanied by a reduction in specific infectivity of >10(5)-fold, to undetectable levels, despite the ability of adapted "protein-only" PrP(Sc) molecules to self-propagate in vitro. We also report that changing only the cofactor component of a minimal reaction substrate mixture during serial propagation induced major changes in the strain properties of an infectious recombinant prion. Moreover, propagation with only one functional cofactor (phosphatidylethanolamine) induced the conversion of three distinct strains into a single strain with unique infectious properties and PrP(Sc) structure. Taken together, these results indicate that cofactor molecules can regulate the defining features of mammalian prions: PrP(Sc) conformation, infectivity, and strain properties. These findings suggest that cofactor molecules likely are integral components of infectious prions.
Conflict of interest statement
Conflict of interest statement: S.S. and N.D. are inventors on a patent held by Dartmouth College on the use of phosphatidylethanolamine as a prion cofactor.
Figures



References
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. - PubMed
-
- Caughey BW, et al. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. - PubMed
-
- Kocisko DA, et al. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

