Calpain 2 activated through N-methyl-D-aspartic acid receptor signaling cleaves CPEB3 and abrogates CPEB3-repressed translation in neurons
- PMID: 22711986
- PMCID: PMC3434545
- DOI: 10.1128/MCB.00296-12
Calpain 2 activated through N-methyl-D-aspartic acid receptor signaling cleaves CPEB3 and abrogates CPEB3-repressed translation in neurons
Abstract
Long-term memory requires the activity-dependent reorganization of the synaptic proteome to modulate synaptic efficacy and consequently consolidate memory. Activity-regulated RNA translation can change the protein composition at the stimulated synapse. Cytoplasmic polyadenylation element-binding protein 3 (CPEB3) is a sequence-specific RNA-binding protein that represses translation of its target mRNAs in neurons, while activation of N-methyl-d-aspartic acid (NMDA) receptors alleviates this repression. Although recent research has revealed the mechanism of CPEB3-inhibited translation, how NMDA receptor signaling modulates the translational activity of CPEB3 remains unclear. This study shows that the repressor CPEB3 is degraded in NMDA-stimulated neurons and that the degradation of CPEB3 is accompanied by the elevated expression of CPEB3's target, epidermal growth factor receptor (EGFR), mostly at the translational level. Using pharmacological and knockdown approaches, we have identified that calpain 2, activated by the influx of calcium through NMDA receptors, proteolyzes the N-terminal repression motif but not the C-terminal RNA-binding domain of CPEB3. As a result, the calpain 2-cleaved CPEB3 fragment binds to RNA but fails to repress translation. Therefore, the cleavage of CPEB3 by NMDA-activated calpain 2 accounts for the activity-related translation of CPEB3-targeted RNAs.
Figures

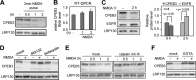

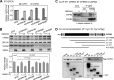




References
-
- Abe K, Takeichi M. 2007. NMDA-receptor activation induces calpain-mediated beta-catenin cleavages for triggering gene expression. Neuron 53:387–397 - PubMed
-
- Adamec E, Beermann ML, Nixon RA. 1998. Calpain I activation in rat hippocampal neurons in culture is NMDA receptor selective and not essential for excitotoxic cell death. Brain Res. Mol. Brain Res. 54:35–48 - PubMed
-
- Banker G, Goslin K. 1988. Developments in neuronal cell culture. Nature 336:185–186 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
