The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo
- PMID: 22711990
- PMCID: PMC3434552
- DOI: 10.1128/MCB.00173-12
The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo
Abstract
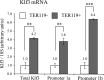
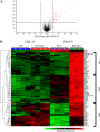
The CACCC-box binding protein erythroid Krüppel-like factor (EKLF/KLF1) is a master regulator that directs the expression of many important erythroid genes. We have previously shown that EKLF drives transcription of the gene for a second KLF, basic Krüppel-like factor, or KLF3. We have now tested the in vivo role of KLF3 in erythroid cells by examining Klf3 knockout mice. KLF3-deficient adults exhibit a mild compensated anemia, including enlarged spleens, increased red pulp, and a higher percentage of erythroid progenitors, together with elevated reticulocytes and abnormal erythrocytes in the peripheral blood. Impaired erythroid maturation is also observed in the fetal liver. We have found that KLF3 levels rise as erythroid cells mature to become TER119(+). Consistent with this, microarray analysis of both TER119(-) and TER119(+) erythroid populations revealed that KLF3 is most critical at the later stages of erythroid maturation and is indeed primarily a transcriptional repressor. Notably, many of the genes repressed by KLF3 are also known to be activated by EKLF. However, the majority of these are not currently recognized as erythroid-cell-specific genes. These results reveal the molecular and physiological function of KLF3, defining it as a feedback repressor that counters the activity of EKLF at selected target genes to achieve normal erythropoiesis.
Figures






References
-
- Coghill E, et al. 2001. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 97:1861–1868 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
