Identification of novel mitosis regulators through data mining with human centromere/kinetochore proteins as group queries
- PMID: 22712476
- PMCID: PMC3419070
- DOI: 10.1186/1471-2121-13-15
Identification of novel mitosis regulators through data mining with human centromere/kinetochore proteins as group queries
Abstract
Background: Proteins functioning in the same biological pathway tend to be transcriptionally co-regulated or form protein-protein interactions (PPI). Multiple spatially and temporally regulated events are coordinated during mitosis to achieve faithful chromosome segregation. The molecular players participating in mitosis regulation are still being unravelled experimentally or using in silico methods.
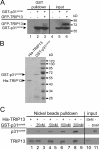
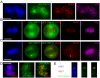
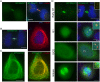
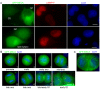
Results: An extensive literature review has led to a compilation of 196 human centromere/kinetochore proteins, all with experimental evidence supporting the subcellular localization. Sixty-four were designated as "core" centromere/kinetochore components based on peak expression and/or well-characterized functions during mitosis. By interrogating and integrating online resources, we have mined for genes/proteins that display transcriptional co-expression or PPI with the core centromere/kinetochore components. Top-ranked hubs in either co-expression or PPI network are not only enriched with known mitosis regulators, but also contain candidates whose mitotic functions are not yet established. Experimental validation found that KIAA1377 is a novel centrosomal protein that also associates with microtubules and midbody; while TRIP13 is a novel kinetochore protein and directly interacts with mitotic checkpoint silencing protein p31(comet).
Conclusions: Transcriptional co-expression and PPI network analyses with known human centromere/kinetochore proteins as a query group help identify novel potential mitosis regulators.
Figures

References
-
- Pines J, Rieder CL. Re-staging mitosis: a contemporary view of mitotic progression. Nature cell biology. 2001;3(1):E3–E6. - PubMed
-
- Muller GA, Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 277(4):877–893. - PubMed
-
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nature cell biology. 2005;7(2):126–136. - PubMed
-
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

