Potential role for PADI-mediated histone citrullination in preimplantation development
- PMID: 22712504
- PMCID: PMC3430579
- DOI: 10.1186/1471-213X-12-19
Potential role for PADI-mediated histone citrullination in preimplantation development
Abstract
Background: The peptidylarginine deiminases (PADIs) convert positively charged arginine residues to neutrally charged citrulline on protein substrates in a process that is known as citrullination or deimination. Previous reports have documented roles for histone citrullination in chromatin remodeling and gene regulation in several tissue types, however, a potential role for histone citrullination in chromatin-based activities during early embryogenesis has not been investigated.
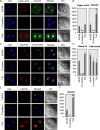
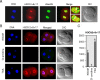
Results: In the present study, we tested by laser scanning confocal indirect immunofluorescence microscopy whether specific arginine residues on the histone H3 and H4 N-terminal tails (H4R3, H3R2 + 8 + 17, and H3R26) were citrullinated in mouse oocytes and preimplantation embryos. Results showed that all of the tested residues were deiminated with each site showing a unique localization pattern during early development. Given these findings, we next tested whether inhibition of PADI activity using the PADI-specific inhibitor, Cl-amidine, may affect embryonic development. We found that treatment of pronuclear stage zygotes with Cl-amidine reduces both histone H3 and H4 tail citrullination and also potently blocks early cleavage divisions in vitro. Additionally, we found that the Cl-amidine treatment reduces acetylation at histone H3K9, H3K18, and H4K5 while having no apparent effect on the repressive histone H3K9 dimethylation modification. Lastly, we found that treatment of zygotes with trichostatin A (TSA) to induce hyperacetylation also resulted in an increase in histone citrullination at H3R2 + 8 + 17.
Conclusions: Given the observed effects of Cl-amidine on embryonic development and the well documented correlation between histone acetylation and transcriptional activation, our findings suggest that histone citrullination may play an important role in facilitating gene expression in early embryos by creating a chromatin environment that is permissive for histone acetylation.
Figures


Similar articles
-
Role of peptidylarginine deiminase 4 (PAD4) in pig parthenogenetic preimplantation embryonic development.Zygote. 2013 Nov;21(4):385-93. doi: 10.1017/S0967199412000160. Epub 2012 Jul 13. Zygote. 2013. PMID: 22793990
-
Localization and expression of peptidylarginine deiminase 4 (PAD4) in mammalian oocytes and preimplantation embryos.Zygote. 2013 Nov;21(4):314-24. doi: 10.1017/S0967199411000633. Epub 2011 Nov 30. Zygote. 2013. PMID: 22126893
-
Citrullination regulates pluripotency and histone H1 binding to chromatin.Nature. 2014 Mar 6;507(7490):104-8. doi: 10.1038/nature12942. Epub 2014 Jan 26. Nature. 2014. PMID: 24463520 Free PMC article.
-
Peptidylarginine deiminase enzymes and citrullinated proteins in female reproductive physiology and associated diseases†.Biol Reprod. 2022 Dec 10;107(6):1395-1410. doi: 10.1093/biolre/ioac173. Biol Reprod. 2022. PMID: 36087287 Free PMC article. Review.
-
Role of citrullination modification catalyzed by peptidylarginine deiminase 4 in gene transcriptional regulation.Acta Biochim Biophys Sin (Shanghai). 2017 Jul 1;49(7):567-572. doi: 10.1093/abbs/gmx042. Acta Biochim Biophys Sin (Shanghai). 2017. PMID: 28472221 Review.
Cited by
-
As the fat flies: The dynamic lipid droplets of Drosophila embryos.Biochim Biophys Acta. 2015 Sep;1851(9):1156-85. doi: 10.1016/j.bbalip.2015.04.002. Epub 2015 Apr 13. Biochim Biophys Acta. 2015. PMID: 25882628 Free PMC article. Review.
-
A Hairy Cituation - PADIs in Regeneration and Alopecia.Front Cell Dev Biol. 2021 Dec 13;9:789676. doi: 10.3389/fcell.2021.789676. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34966743 Free PMC article. Review.
-
Developmentally regulated H2Av buffering via dynamic sequestration to lipid droplets in Drosophila embryos.Elife. 2018 Jul 25;7:e36021. doi: 10.7554/eLife.36021. Elife. 2018. PMID: 30044219 Free PMC article.
-
PADI6: What we know about the elusive fifth member of the peptidyl arginine deiminase family.Philos Trans R Soc Lond B Biol Sci. 2023 Nov 20;378(1890):20220242. doi: 10.1098/rstb.2022.0242. Epub 2023 Oct 2. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 37778376 Free PMC article. Review.
-
Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei.Sci Rep. 2021 May 12;11(1):10146. doi: 10.1038/s41598-021-89334-w. Sci Rep. 2021. PMID: 33980975 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

