Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans
- PMID: 22712530
- PMCID: PMC3443458
- DOI: 10.1186/1471-2164-13-254
Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans
Abstract
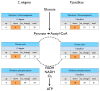
Background: An organism can respond to changing environmental conditions by adjusting gene regulation and by forming alternative phenotypes. In nematodes, these mechanisms are coupled because many species will form dauer larvae, a stress-resistant and non-aging developmental stage, when exposed to unfavorable environmental conditions, and execute gene expression programs that have been selected for the survival of the animal in the wild. These dauer larvae represent an environmentally induced, homologous developmental stage across many nematode species, sharing conserved morphological and physiological properties. Hence it can be expected that some core components of the associated transcriptional program would be conserved across species, while others might diverge over the course of evolution. However, transcriptional and metabolic analysis of dauer development has been largely restricted to Caenorhabditis elegans. Here, we use a transcriptomic approach to compare the dauer stage in the evolutionary model system Pristionchus pacificus with the dauer stage in C. elegans.
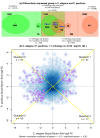
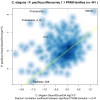
Results: We have employed Agilent microarrays, which represent 20,446 P. pacificus and 20,143 C. elegans genes to show an unexpected divergence in the expression profiles of these two nematodes in dauer and dauer exit samples. P. pacificus and C. elegans differ in the dynamics and function of genes that are differentially expressed. We find that only a small number of orthologous gene pairs show similar expression pattern in the dauers of the two species, while the non-orthologous fraction of genes is a major contributor to the active transcriptome in dauers. Interestingly, many of the genes acquired by horizontal gene transfer and orphan genes in P. pacificus, are differentially expressed suggesting that these genes are of evolutionary and functional importance.
Conclusion: Our data set provides a catalog for future functional investigations and indicates novel insight into evolutionary mechanisms. We discuss the limited conservation of core developmental and transcriptional programs as a common aspect of animal evolution.
Figures
References
-
- Fuchs G. Die Naturgeschichte der Nematoden und einiger anderer Parasiten 1. des Ips typographus L. 2. des Hylobius abietis L. Zool Jahrb. 1915;38:109–222.
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

