Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage
- PMID: 22712728
- PMCID: PMC7659207
- DOI: 10.1111/j.1349-7006.2012.02362.x
Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage
Abstract
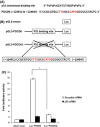
The cellular response to genotoxic stress is multifaceted in nature. Following DNA damage, the tumor suppressor gene p53 activates and plays critical roles in cell cycle arrest, activation of DNA repair and in the event of irreparable damage, induction of apoptosis. The breakdown of apoptosis causes the accumulation of mutant cells. The elucidation of the mechanism for the p53-dependent apoptosis will be crucial in applying the strategy for cancer patients. However, the mechanism of p53-dependent apoptosis remains largely unclear. Here, we carried out ChIP followed by massively parallel DNA sequencing assay (ChIP-seq) to uncover mechanisms of apoptosis. Using ChIP-seq, we identified PDCD6 as a novel p53-responsive gene. We determined putative p53-binding sites that are important for p53 regulation in response to DNA damage in the promoter region of PDCD6. Knockdown of PDCD6 suppressed p53-dependent apoptosis. We also observed that cytochrome c release and the cleavage of PARP by caspase-3 were suppressed by depletion of PDCD6. We further observed that PDCD6 localizes in the nucleus in response to DNA damage. We identified the nuclear localization signal of PDCD6 and, importantly, the nuclear accumulation of PDCD6 significantly induced apoptosis after genotoxic stress. Therefore, we conclude that a novel p53-responsive gene PDCD6 is accumulated in the nucleus and induces apoptosis in response to DNA damage.
© 2012 Japanese Cancer Association.
Figures




References
-
- Birch JM, Blair V, Kelsey AM et al Cancer phenotype correlates with constitutional TP53 genotype in families with the Li‐Fraumeni syndrome. Oncogene 1998; 17: 1061–8. - PubMed
-
- Olivier M, Goldgar DE, Sodha N et al Li‐Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res 2003; 63: 6643–50. - PubMed
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med 2004; 10: 789–99. - PubMed
-
- Finlay CA, Hinds PW, Levine AJ. The p53 proto‐oncogene can act as a suppressor of transformation. Cell 1989; 57: 1083–93. - PubMed
-
- Sherr CJ. Principles of tumor suppression. Cell 2004; 116: 235–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

