A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase
- PMID: 22713238
- PMCID: PMC3404880
- DOI: 10.4161/cc.20919
A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase
Abstract
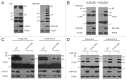
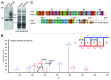
Ubiquitination of histones plays a critical role in the regulation of several processes within the nucleus, including maintenance of genome stability and transcriptional regulation. The only known ubiquitination site on histones is represented by a conserved Lys residue located at the C terminus of the protein. Here, we describe a novel ubiquitin mark at the N-terminal tail of histone H2As consisting of two Lys residues at positions 13 and 15 (K13/K15). This "bidentate" site is a target of the DNA damage response (DDR) ubiquitin ligases RNF8 and RNF168. Histone mutants lacking the K13/K15 site impair RNF168- and DNA damage-dependent ubiquitination. Conversely, inactivation of the canonical C-terminal site prevents the constitutive monoubiquitination of histone H2As but does not abolish the ubiquitination induced by RNF168. A ubiquitination-defective mutant is obtained by inactivating both the N- and the C-terminal sites, suggesting that these are unique, non-redundant acceptors of ubiquitination on histone H2As. This unprecedented result implies that RNF168 generates a qualitatively different Ub mark on chromatin.
Figures

Comment in
-
Ubiquitin and the DNA damage response: a new handle on histones.Cell Cycle. 2012 Sep 1;11(17):3153. doi: 10.4161/cc.21718. Epub 2012 Aug 16. Cell Cycle. 2012. PMID: 22895171 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
