Effector CD4+ T cell expression signatures and immune-mediated disease associated genes
- PMID: 22715389
- PMCID: PMC3371029
- DOI: 10.1371/journal.pone.0038510
Effector CD4+ T cell expression signatures and immune-mediated disease associated genes
Abstract
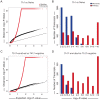
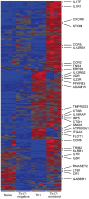
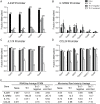
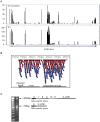
Genome-wide association studies (GWAS) in immune-mediated diseases have identified over 150 associated genomic loci. Many of these loci play a role in T cell responses, and regulation of T cell differentiation plays a critical role in immune-mediated diseases; however, the relationship between implicated disease loci and T cell differentiation is incompletely understood. To further address this relationship, we examined differential gene expression in naïve human CD4+ T cells, as well as in in vitro differentiated Th1, memory Th17-negative and Th17-enriched CD4+ T cells subsets using microarray and RNASeq. We observed a marked enrichment for increased expression in memory CD4+ compared to naïve CD4+ T cells of genes contained among immune-mediated disease loci. Within memory T cells, expression of disease-associated genes was typically increased in Th17-enriched compared to Th17-negative cells. Utilizing RNASeq and promoter methylation studies, we identified a differential regulation pattern for genes solely expressed in Th17 cells (IL17A and CCL20) compared to genes expressed in both Th17 and Th1 cells (IL23R and IL12RB2), where high levels of promoter methylation are correlated to near zero RNASeq levels for IL17A and CCL20. These findings have implications for human Th17 celI plasticity and for the regulation of Th17-Th1 expression signatures. Importantly, utilizing RNASeq we found an abundant isoform of IL23R terminating before the transmembrane domain that was enriched in Th17 cells. In addition to molecular resolution, we find that RNASeq provides significantly improved power to define differential gene expression and identify alternative gene variants relative to microarray analysis. The comprehensive integration of differential gene expression between cell subsets with disease-association signals, and functional pathways provides insight into disease pathogenesis.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

