Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking
- PMID: 22716043
- PMCID: PMC3405771
- DOI: 10.1021/jm300687e
Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking
Abstract
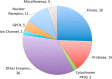
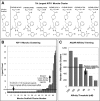
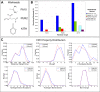
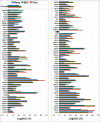
A key metric to assess molecular docking remains ligand enrichment against challenging decoys. Whereas the directory of useful decoys (DUD) has been widely used, clear areas for optimization have emerged. Here we describe an improved benchmarking set that includes more diverse targets such as GPCRs and ion channels, totaling 102 proteins with 22886 clustered ligands drawn from ChEMBL, each with 50 property-matched decoys drawn from ZINC. To ensure chemotype diversity, we cluster each target's ligands by their Bemis-Murcko atomic frameworks. We add net charge to the matched physicochemical properties and include only the most dissimilar decoys, by topology, from the ligands. An online automated tool (http://decoys.docking.org) generates these improved matched decoys for user-supplied ligands. We test this data set by docking all 102 targets, using the results to improve the balance between ligand desolvation and electrostatics in DOCK 3.6. The complete DUD-E benchmarking set is freely available at http://dude.docking.org.
Figures


References
-
- Kitchen D. B.; Decornez H.; Furr J. R.; Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nature Rev. Drug Discovery 2004, 3, 935–949. - PubMed
-
- Gruneberg S.; Stubbs M. T.; Klebe G. Successful virtual screening for novel inhibitors of human carbonic anhydrase: strategy and experimental confirmation. J. Med. Chem. 2002, 45, 3588–3602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

