Molecular evolution of the polyamine oxidase gene family in Metazoa
- PMID: 22716069
- PMCID: PMC3517346
- DOI: 10.1186/1471-2148-12-90
Molecular evolution of the polyamine oxidase gene family in Metazoa
Abstract
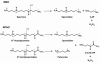
Background: Polyamine oxidase enzymes catalyze the oxidation of polyamines and acetylpolyamines. Since polyamines are basic regulators of cell growth and proliferation, their homeostasis is crucial for cell life. Members of the polyamine oxidase gene family have been identified in a wide variety of animals, including vertebrates, arthropodes, nematodes, placozoa, as well as in plants and fungi. Polyamine oxidases (PAOs) from yeast can oxidize spermine, N1-acetylspermine, and N1-acetylspermidine, however, in vertebrates two different enzymes, namely spermine oxidase (SMO) and acetylpolyamine oxidase (APAO), specifically catalyze the oxidation of spermine, and N1-acetylspermine/N1-acetylspermidine, respectively. Little is known about the molecular evolutionary history of these enzymes. However, since the yeast PAO is able to catalyze the oxidation of both acetylated and non acetylated polyamines, and in vertebrates these functions are addressed by two specialized polyamine oxidase subfamilies (APAO and SMO), it can be hypothesized an ancestral reference for the former enzyme from which the latter would have been derived.
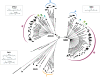
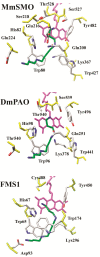
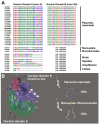
Results: We analysed 36 SMO, 26 APAO, and 14 PAO homologue protein sequences from 54 taxa including various vertebrates and invertebrates. The analysis of the full-length sequences and the principal domains of vertebrate and invertebrate PAOs yielded consensus primary protein sequences for vertebrate SMOs and APAOs, and invertebrate PAOs. This analysis, coupled to molecular modeling techniques, also unveiled sequence regions that confer specific structural and functional properties, including substrate specificity, by the different PAO subfamilies. Molecular phylogenetic trees revealed a basal position of all the invertebrates PAO enzymes relative to vertebrate SMOs and APAOs. PAOs from insects constitute a monophyletic clade. Two PAO variants sampled in the amphioxus are basal to the dichotomy between two well supported monophyletic clades including, respectively, all the SMOs and APAOs from vertebrates. The two vertebrate monophyletic clades clustered strictly mirroring the organismal phylogeny of fishes, amphibians, reptiles, birds, and mammals. Evidences from comparative genomic analysis, structural evolution and functional divergence in a phylogenetic framework across Metazoa suggested an evolutionary scenario where the ancestor PAO coding sequence, present in invertebrates as an orthologous gene, has been duplicated in the vertebrate branch to originate the paralogous SMO and APAO genes. A further genome evolution event concerns the SMO gene of placental, but not marsupial and monotremate, mammals which increased its functional variation following an alternative splicing (AS) mechanism.
Conclusions: In this study the explicit integration in a phylogenomic framework of phylogenetic tree construction, structure prediction, and biochemical function data/prediction, allowed inferring the molecular evolutionary history of the PAO gene family and to disambiguate paralogous genes related by duplication event (SMO and APAO) and orthologous genes related by speciation events (PAOs, SMOs/APAOs). Further, while in vertebrates experimental data corroborate SMO and APAO molecular function predictions, in invertebrates the finding of a supported phylogenetic clusters of insect PAOs and the co-occurrence of two PAO variants in the amphioxus urgently claim the need for future structure-function studies.
Figures

Similar articles
-
Identification and biochemical characterization of polyamine oxidases in amphioxus: Implications for emergence of vertebrate-specific spermine and acetylpolyamine oxidases.Gene. 2016 Jan 10;575(2 Pt 2):429-437. doi: 10.1016/j.gene.2015.09.017. Epub 2015 Sep 12. Gene. 2016. PMID: 26367330
-
Pacific oyster polyamine oxidase: a protein missing link in invertebrate evolution.Amino Acids. 2015 May;47(5):949-61. doi: 10.1007/s00726-015-1924-2. Epub 2015 Feb 6. Amino Acids. 2015. PMID: 25655384
-
Genomic identification and biochemical characterization of the mammalian polyamine oxidase involved in polyamine back-conversion.Biochem J. 2003 Feb 15;370(Pt 1):19-28. doi: 10.1042/BJ20021779. Biochem J. 2003. PMID: 12477380 Free PMC article.
-
Polyamine oxidase, properties and functions.Prog Brain Res. 1995;106:333-44. doi: 10.1016/s0079-6123(08)61229-7. Prog Brain Res. 1995. PMID: 8584670 Review.
-
Structure-function relationships in the evolutionary framework of spermine oxidase.J Mol Evol. 2013 Jun;76(6):365-70. doi: 10.1007/s00239-013-9570-3. Epub 2013 Jul 5. J Mol Evol. 2013. PMID: 23828398 Review.
Cited by
-
The tree of life of polyamine oxidases.Sci Rep. 2020 Oct 20;10(1):17858. doi: 10.1038/s41598-020-74708-3. Sci Rep. 2020. PMID: 33082384 Free PMC article.
-
The Involvement of Polyamines Catabolism in the Crosstalk between Neurons and Astrocytes in Neurodegeneration.Biomedicines. 2022 Jul 21;10(7):1756. doi: 10.3390/biomedicines10071756. Biomedicines. 2022. PMID: 35885061 Free PMC article. Review.
-
Metabolomic Reprogramming of C57BL/6-Macrophages during Early Infection with L. amazonensis.Int J Mol Sci. 2021 Jun 26;22(13):6883. doi: 10.3390/ijms22136883. Int J Mol Sci. 2021. PMID: 34206906 Free PMC article.
-
Transcriptome and co-expression network analysis reveals the molecular mechanism of inosine monophosphate-specific deposition in chicken muscle.Front Physiol. 2023 May 17;14:1199311. doi: 10.3389/fphys.2023.1199311. eCollection 2023. Front Physiol. 2023. PMID: 37265843 Free PMC article.
-
Polyamine Oxidases Play Various Roles in Plant Development and Abiotic Stress Tolerance.Plants (Basel). 2019 Jun 21;8(6):184. doi: 10.3390/plants8060184. Plants (Basel). 2019. PMID: 31234345 Free PMC article. Review.
References
-
- Cohen SS. In: A guide to the polyamines. Cohen SS, editor. Oxford University Press, New York; 1998. Polyamine oxidases and dehydrogenases; pp. 1–82.
-
- Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA Jr. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61:5370–5373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

