Phylogenetic relationship of dengue virus type 3 isolated in Brazil and Paraguay and global evolutionary divergence dynamics
- PMID: 22716071
- PMCID: PMC3494512
- DOI: 10.1186/1743-422X-9-124
Phylogenetic relationship of dengue virus type 3 isolated in Brazil and Paraguay and global evolutionary divergence dynamics
Abstract
Background: Dengue is the most important mosquito-borne viral disease worldwide. Dengue virus comprises four antigenically related viruses named dengue virus type 1 to 4 (DENV1-4). DENV-3 was re-introduced into the Americas in 1994 causing outbreaks in Nicaragua and Panama. DENV-3 was introduced in Brazil in 2000 and then spread to most of the Brazilian States, reaching the neighboring country, Paraguay in 2002. In this study, we have analyzed the phylogenetic relationship of DENV-3 isolated in Brazil and Paraguay with viruses isolated worldwide. We have also analyzed the evolutionary divergence dynamics of DENV-3 viruses.
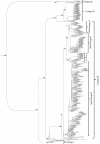
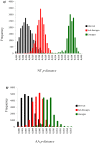
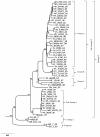
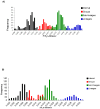
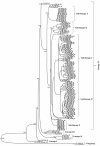
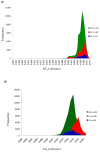
Results: The entire open reading frame (ORF) of thirteen DENV-3 isolated in Brazil (n = 9) and Paraguay (n = 4) were sequenced for phylogenetic analysis. DENV-3 grouped into three main genotypes (I, II and III). Several internal clades were found within each genotype that we called lineage and sub-lineage. Viruses included in this study belong to genotype III and grouped together with viruses isolated in the Americas within the lineage III. The Brazilian viruses were further segregated into two different sub-lineage, A and B, and the Paraguayan into the sub-lineage B. All three genotypes showed internal grouping. The nucleotide divergence was in average 6.7% for genotypes, 2.7% for lineages and 1.5% for sub-lineages. Phylogenetic trees constructed with any of the protein gene sequences showed the same segregation of the DENV-3 in three genotypes.
Conclusion: Our results showed that two groups of DENV-3 genotypes III circulated in Brazil during 2002-2009, suggesting different events of introduction of the virus through different regions of the country. In Paraguay, only one group DENV-3 genotype III is circulating that is very closely related to the Brazilian viruses of sub-lineage B. Different degree of grouping can be observed for DENV-3 and each group showed a characteristic evolutionary divergence. Finally, we have observed that any protein gene sequence can be used to identify the virus genotype.
Figures


References
-
- WHO. Dengue: guidelines for diagnosis, treatment, prevention and control. Geneva: World Health Organization; 2009. - PubMed
-
- CDC. From the Centers for Disease Control and Prevention. Dengue type 3 infection--Nicaragua and Panama, October-November 1994. JAMA. 1995;273:840–841. - PubMed
-
- Guzmán MG, Vázquez S, Martínez E, Alvarez M, Rodríguez R, Kourí G, de los Reyes J, Acevedo F. [Dengue in Nicaragua, 1994: reintroduction of serotype 3 in the Americas] Bol Oficina Sanit Panam. 1996;121:102–110. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

