Evidence for roles of the Escherichia coli Hda protein beyond regulatory inactivation of DnaA
- PMID: 22716942
- PMCID: PMC3418461
- DOI: 10.1111/j.1365-2958.2012.08129.x
Evidence for roles of the Escherichia coli Hda protein beyond regulatory inactivation of DnaA
Abstract
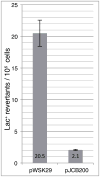
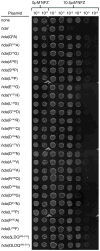
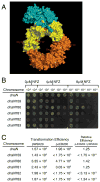
The ATP-bound form of the Escherichia coli DnaA protein binds 'DnaA boxes' present in the origin of replication (oriC) and operator sites of several genes, including dnaA, to co-ordinate their transcription with initiation of replication. The Hda protein, together with the β sliding clamp, stimulates the ATPase activity of DnaA via a process termed regulatory inactivation of DnaA (RIDA), to regulate the activity of DnaA in DNA replication. Here, we used the mutant dnaN159 strain, which expresses the β159 clamp protein, to gain insight into how the actions of Hda are co-ordinated with replication. Elevated expression of Hda impeded growth of the dnaN159 strain in a Pol II- and Pol IV-dependent manner, suggesting a role for Hda managing the actions of these Pols. In a wild-type strain, elevated levels of Hda conferred sensitivity to nitrofurazone, and suppressed the frequency of -1 frameshift mutations characteristic of Pol IV, while loss of hda conferred cold sensitivity. Using the dnaN159 strain, we identified 24 novel hda alleles, four of which supported E. coli viability despite their RIDA defect. Taken together, these findings suggest that although one or more Hda functions are essential for cell viability, RIDA may be dispensable.
© 2012 Blackwell Publishing Ltd.
Figures


References
-
- Atlung T, Lobner-Olesen A, Hansen FG. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Molecular & general genetics: MGG. 1987;206:51–59. - PubMed
-
- Boye E, Blinkova A, Walker JR. Defective initiation in an Escherichia coli dnaA(Cs,Sx) mutant. Biochimie. 2001;83:25–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

