Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs
- PMID: 22718037
- PMCID: PMC3392844
- DOI: 10.1136/amiajnl-2011-000699
Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs
Abstract
Objective: Adverse drug reaction (ADR) is one of the major causes of failure in drug development. Severe ADRs that go undetected until the post-marketing phase of a drug often lead to patient morbidity. Accurate prediction of potential ADRs is required in the entire life cycle of a drug, including early stages of drug design, different phases of clinical trials, and post-marketing surveillance.
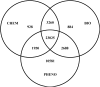
Methods: Many studies have utilized either chemical structures or molecular pathways of the drugs to predict ADRs. Here, the authors propose a machine-learning-based approach for ADR prediction by integrating the phenotypic characteristics of a drug, including indications and other known ADRs, with the drug's chemical structures and biological properties, including protein targets and pathway information. A large-scale study was conducted to predict 1385 known ADRs of 832 approved drugs, and five machine-learning algorithms for this task were compared.
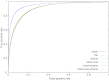
Results: This evaluation, based on a fivefold cross-validation, showed that the support vector machine algorithm outperformed the others. Of the three types of information, phenotypic data were the most informative for ADR prediction. When biological and phenotypic features were added to the baseline chemical information, the ADR prediction model achieved significant improvements in area under the curve (from 0.9054 to 0.9524), precision (from 43.37% to 66.17%), and recall (from 49.25% to 63.06%). Most importantly, the proposed model successfully predicted the ADRs associated with withdrawal of rofecoxib and cerivastatin.
Conclusion: The results suggest that phenotypic information on drugs is valuable for ADR prediction. Moreover, they demonstrate that different models that combine chemical, biological, or phenotypic information can be built from approved drugs, and they have the potential to detect clinically important ADRs in both preclinical and post-marketing phases.
Conflict of interest statement
Figures



References
-
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998;279:1200–5 - PubMed
-
- Moore TJ, Cohen MR, Furberg CD. Serious adverse drug events reported to the Food and Drug Administration, 1998–2005. Arch Intern Med 2007;167:1752–9 - PubMed
-
- Giacomini KM, Krauss RM, Roden DM, et al. When good drugs go bad. Nature 2007;446:975–7 - PubMed
-
- Whitebread S, Hamon J, Bojanic D, et al. Keynote review: in vitro safety pharmacology profiling: an essential tool for successful drug development. Drug Discov Today 2005;10:1421–33 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

