Control of the mitotic exit network during meiosis
- PMID: 22718910
- PMCID: PMC3418307
- DOI: 10.1091/mbc.E12-03-0235
Control of the mitotic exit network during meiosis
Abstract
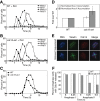
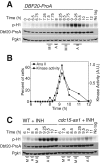
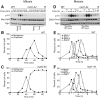
The mitotic exit network (MEN) is an essential GTPase signaling pathway that triggers exit from mitosis in budding yeast. We show here that during meiosis, the MEN is dispensable for exit from meiosis I but contributes to the timely exit from meiosis II. Consistent with a role for the MEN during meiosis II, we find that the signaling pathway is active only during meiosis II. Our analysis further shows that MEN signaling is modulated during meiosis in several key ways. Whereas binding of MEN components to spindle pole bodies (SPBs) is necessary for MEN signaling during mitosis, during meiosis MEN signaling occurs off SPBs and does not require the SPB recruitment factor Nud1. Furthermore, unlike during mitosis, MEN signaling is controlled through the regulated interaction between the MEN kinase Dbf20 and its activating subunit Mob1. Our data lead to the conclusion that a pathway essential for vegetative growth is largely dispensable for the specialized meiotic divisions and provide insights into how cell cycle regulatory pathways are modulated to accommodate different modes of cell division.
Figures



Similar articles
-
The Mitotic Exit Network Regulates Spindle Pole Body Selection During Sporulation of Saccharomyces cerevisiae.Genetics. 2017 Jun;206(2):919-937. doi: 10.1534/genetics.116.194522. Epub 2017 Apr 26. Genetics. 2017. PMID: 28450458 Free PMC article.
-
Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes.Science. 2013 May 17;340(6134):871-5. doi: 10.1126/science.1235822. Epub 2013 Apr 11. Science. 2013. PMID: 23579499 Free PMC article.
-
Budding yeast Wee1 distinguishes spindle pole bodies to guide their pattern of age-dependent segregation.Nat Cell Biol. 2017 Aug;19(8):941-951. doi: 10.1038/ncb3576. Epub 2017 Jul 17. Nat Cell Biol. 2017. PMID: 28714971
-
Mitotic exit control: a space and time odyssey.Curr Biol. 2011 Oct 25;21(20):R857-9. doi: 10.1016/j.cub.2011.09.023. Curr Biol. 2011. PMID: 22032192 Review.
-
Regulation of Mitotic Exit in Saccharomyces cerevisiae.Methods Mol Biol. 2017;1505:3-17. doi: 10.1007/978-1-4939-6502-1_1. Methods Mol Biol. 2017. PMID: 27826852 Review.
Cited by
-
The Mitotic Exit Network Regulates Spindle Pole Body Selection During Sporulation of Saccharomyces cerevisiae.Genetics. 2017 Jun;206(2):919-937. doi: 10.1534/genetics.116.194522. Epub 2017 Apr 26. Genetics. 2017. PMID: 28450458 Free PMC article.
-
A Noncanonical Hippo Pathway Regulates Spindle Disassembly and Cytokinesis During Meiosis in Saccharomyces cerevisiae.Genetics. 2020 Oct;216(2):447-462. doi: 10.1534/genetics.120.303584. Epub 2020 Aug 11. Genetics. 2020. PMID: 32788308 Free PMC article.
-
Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast.Mol Biol Cell. 2015 Apr 15;26(8):1440-51. doi: 10.1091/mbc.E14-10-1442. Epub 2015 Feb 18. Mol Biol Cell. 2015. PMID: 25694455 Free PMC article.
-
Meiosis II spindle disassembly requires two distinct pathways.Mol Biol Cell. 2023 Sep 1;34(10):ar98. doi: 10.1091/mbc.E23-03-0096. Epub 2023 Jul 12. Mol Biol Cell. 2023. PMID: 37436806 Free PMC article.
-
The DNA damage checkpoint and the spindle position checkpoint: guardians of meiotic commitment.Curr Genet. 2019 Oct;65(5):1135-1140. doi: 10.1007/s00294-019-00981-z. Epub 2019 Apr 26. Curr Genet. 2019. PMID: 31028453 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

