Analysis of rRNA processing and translation in mammalian cells using a synthetic 18S rRNA expression system
- PMID: 22718970
- PMCID: PMC3439915
- DOI: 10.1093/nar/gks530
Analysis of rRNA processing and translation in mammalian cells using a synthetic 18S rRNA expression system
Abstract
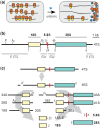
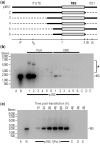
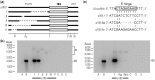
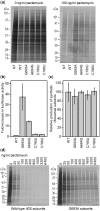
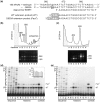
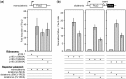
Analysis of processing, assembly, and function of higher eukaryotic ribosomal RNA (rRNA) has been hindered by the lack of an expression system that enables rRNA to be modified and then examined functionally. Given the potential usefulness of such a system, we have developed one for mammalian 18S rRNA. We inserted a sequence tag into expansion segment 3 of mouse 18S rRNA to monitor expression and cleavage by hybridization. Mutations were identified that confer resistance to pactamycin, allowing functional analysis of 40S ribosomal subunits containing synthetic 18S rRNAs by selectively blocking translation from endogenous (pactamycin-sensitive) subunits. rRNA constructs were suitably expressed in transfected cells, shown to process correctly, incorporate into ≈ 15% of 40S subunits, and function normally based on various criteria. After rigorous analysis, the system was used to investigate the importance of sequences that flank 18S rRNA in precursor transcripts. Although deletion analysis supported the requirement of binding sites for the U3 snoRNA, it showed that a large segment of the 5' external transcribed spacer and the entire first internal transcribed spacer, both of which flank 18S rRNA, are not required. The success of this approach opens the possibility of functional analyses of ribosomes, with applications in basic research and synthetic biology.
Figures
Similar articles
-
hUTP24 is essential for processing of the human rRNA precursor at site A1, but not at site A0.RNA Biol. 2015;12(9):1010-29. doi: 10.1080/15476286.2015.1073437. RNA Biol. 2015. PMID: 26237581 Free PMC article.
-
Imp3 unfolds stem structures in pre-rRNA and U3 snoRNA to form a duplex essential for small subunit processing.RNA. 2013 Oct;19(10):1372-83. doi: 10.1261/rna.039511.113. Epub 2013 Aug 26. RNA. 2013. PMID: 23980203 Free PMC article.
-
Base pairing between U3 small nucleolar RNA and the 5' end of 18S rRNA is required for pre-rRNA processing.Mol Cell Biol. 1999 Sep;19(9):6012-9. doi: 10.1128/MCB.19.9.6012. Mol Cell Biol. 1999. PMID: 10454548 Free PMC article.
-
Processing of mammalian rRNA precursors at the 3' end of 18S rRNA. Identification of cis-acting signals suggests the involvement of U13 small nucleolar RNA.Eur J Biochem. 1996 Dec 1;242(2):206-13. doi: 10.1111/j.1432-1033.1996.0206r.x. Eur J Biochem. 1996. PMID: 8973634
-
Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors.Nucleic Acids Symp Ser. 1997;(36):64-7. Nucleic Acids Symp Ser. 1997. PMID: 9478208 Review.
Cited by
-
DLK-Dependent Biphasic Reactivation of Herpes Simplex Virus Latency Established in the Absence of Antivirals.J Virol. 2022 Jun 22;96(12):e0050822. doi: 10.1128/jvi.00508-22. Epub 2022 May 24. J Virol. 2022. PMID: 35608347 Free PMC article.
-
Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo.Proc Natl Acad Sci U S A. 2014 Oct 28;111(43):15385-9. doi: 10.1073/pnas.1413472111. Epub 2014 Oct 13. Proc Natl Acad Sci U S A. 2014. PMID: 25313046 Free PMC article.
-
Translation regulation by ribosomes: Increased complexity and expanded scope.RNA Biol. 2016 Sep;13(9):748-55. doi: 10.1080/15476286.2015.1107701. Epub 2015 Oct 29. RNA Biol. 2016. PMID: 26513496 Free PMC article. Review.
-
Investigating the effect of positional variation on mid-lactation mammary gland transcriptomics in mice fed either a low-fat or high-fat diet.PLoS One. 2021 Aug 26;16(8):e0255770. doi: 10.1371/journal.pone.0255770. eCollection 2021. PLoS One. 2021. PMID: 34437559 Free PMC article.
-
Evaluation of synthetic mRNA with selected UTR sequences and alternative poly(A) tail, in vitro and in vivo.Mol Ther Nucleic Acids. 2025 Jul 30;36(3):102648. doi: 10.1016/j.omtn.2025.102648. eCollection 2025 Sep 9. Mol Ther Nucleic Acids. 2025. PMID: 40822034 Free PMC article.
References
-
- Stelzl U, Connell S, Wittman-Liebold B. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; 2001. Ribosomal proteins: role in ribosomal functions; pp. 1–12.
-
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends. Biochem. Sci. 1996;21:164–165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

