How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes
- PMID: 22718982
- PMCID: PMC3439924
- DOI: 10.1093/nar/gks548
How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes
Abstract
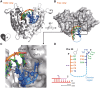
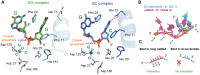
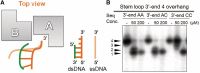
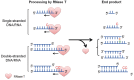
Exonucleases are key enzymes in the maintenance of genome stability, processing of immature RNA precursors and degradation of unnecessary nucleic acids. However, it remains unclear how exonucleases digest nucleic acids to generate correct end products for next-step processing. Here we show how the exonuclease RNase T stops its trimming precisely. The crystal structures of RNase T in complex with a stem-loop DNA, a GG dinucleotide and single-stranded DNA with different 3'-end sequences demonstrate why a duplex with a short 3'-overhang, a dinucleotide and a ssDNA with a 3'-end C cannot be further digested by RNase T. Several hydrophobic residues in RNase T change their conformation upon substrate binding and induce an active or inactive conformation in the active site that construct a precise machine to determine which substrate should be digested based on its sequence, length and structure. These studies thus provide mechanistic insights into how RNase T prevents over digestion of its various substrates, and the results can be extrapolated to the thousands of members of the DEDDh family of exonucleases.
Figures


References
-
- Shevelev IV, Hubscher U. The 3′-5′ exonucleases. Nat. Rev. Mol. Cell. Biol. 2002;3:364–376. - PubMed
-
- Uhrhammer NA, Lafarge L, Dos Santos L, Domaszewska A, Lange M, Yang Y, Aractingi S, Bessis D, Bignon YJ. Werner syndrome and mutations of the WRN and LMNA genes in France. Hum. Mutat. 2006;27:718–719. - PubMed
-
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

