Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells
- PMID: 22719263
- PMCID: PMC3375231
- DOI: 10.1371/journal.pgen.1002749
Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells
Abstract
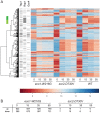
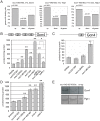
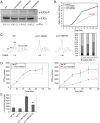
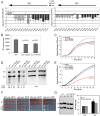
Cohesin is a protein complex known for its essential role in chromosome segregation. However, cohesin and associated factors have additional functions in transcription, DNA damage repair, and chromosome condensation. The human cohesinopathy diseases are thought to stem not from defects in chromosome segregation but from gene expression. The role of cohesin in gene expression is not well understood. We used budding yeast strains bearing mutations analogous to the human cohesinopathy disease alleles under control of their native promoter to study gene expression. These mutations do not significantly affect chromosome segregation. Transcriptional profiling reveals that many targets of the transcriptional activator Gcn4 are induced in the eco1-W216G mutant background. The upregulation of Gcn4 was observed in many cohesin mutants, and this observation suggested protein translation was reduced. We demonstrate that the cohesinopathy mutations eco1-W216G and smc1-Q843Δ are associated with defects in ribosome biogenesis and a reduction in the actively translating fraction of ribosomes, eiF2α-phosphorylation, and (35)S-methionine incorporation, all of which indicate a deficit in protein translation. Metabolic labeling shows that the eco1-W216G and smc1-Q843Δ mutants produce less ribosomal RNA, which is expected to constrain ribosome biogenesis. Further analysis shows that the production of rRNA from an individual repeat is reduced while copy number remains unchanged. Similar defects in rRNA production and protein translation are observed in a human Roberts syndrome cell line. In addition, cohesion is defective specifically at the rDNA locus in the eco1-W216G mutant, as has been previously reported for Roberts syndrome. Collectively, our data suggest that cohesin proteins normally facilitate production of ribosomal RNA and protein translation, and this is one way they can influence gene expression. Reduced translational capacity could contribute to the human cohesinopathies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. - PubMed
-
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. - PubMed
-
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. - PubMed
-
- Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. - PubMed
-
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

