Polymorphisms in the mitochondrial ribosome recycling factor EF-G2mt/MEF2 compromise cell respiratory function and increase atorvastatin toxicity
- PMID: 22719265
- PMCID: PMC3375252
- DOI: 10.1371/journal.pgen.1002755
Polymorphisms in the mitochondrial ribosome recycling factor EF-G2mt/MEF2 compromise cell respiratory function and increase atorvastatin toxicity
Abstract
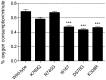
Mitochondrial translation, essential for synthesis of the electron transport chain complexes in the mitochondria, is governed by nuclear encoded genes. Polymorphisms within these genes are increasingly being implicated in disease and may also trigger adverse drug reactions. Statins, a class of HMG-CoA reductase inhibitors used to treat hypercholesterolemia, are among the most widely prescribed drugs in the world. However, a significant proportion of users suffer side effects of varying severity that commonly affect skeletal muscle. The mitochondria are one of the molecular targets of statins, and these drugs have been known to uncover otherwise silent mitochondrial mutations. Based on yeast genetic studies, we identify the mitochondrial translation factor MEF2 as a mediator of atorvastatin toxicity. The human ortholog of MEF2 is the Elongation Factor Gene (EF-G) 2, which has previously been shown to play a specific role in mitochondrial ribosome recycling. Using small interfering RNA (siRNA) silencing of expression in human cell lines, we demonstrate that the EF-G2mt gene is required for cell growth on galactose medium, signifying an essential role for this gene in aerobic respiration. Furthermore, EF-G2mt silenced cell lines have increased susceptibility to cell death in the presence of atorvastatin. Using yeast as a model, conserved amino acid variants, which arise from non-synonymous single nucleotide polymorphisms (SNPs) in the EF-G2mt gene, were generated in the yeast MEF2 gene. Although these mutations do not produce an obvious growth phenotype, three mutations reveal an atorvastatin-sensitive phenotype and further analysis uncovers a decreased respiratory capacity. These findings constitute the first reported phenotype associated with SNPs in the EF-G2mt gene and implicate the human EF-G2mt gene as a pharmacogenetic candidate gene for statin toxicity in humans.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis.Mol Cell. 2009 Aug 28;35(4):502-10. doi: 10.1016/j.molcel.2009.06.028. Mol Cell. 2009. PMID: 19716793
-
The MEF2 gene is essential for yeast longevity, with a dual role in cell respiration and maintenance of mitochondrial membrane potential.FEBS Lett. 2011 Apr 20;585(8):1140-6. doi: 10.1016/j.febslet.2011.03.022. Epub 2011 Mar 21. FEBS Lett. 2011. PMID: 21414316
-
Mitochondrial translation factors of Trypanosoma brucei: elongation factor-Tu has a unique subdomain that is essential for its function.Mol Microbiol. 2013 Nov;90(4):744-55. doi: 10.1111/mmi.12397. Epub 2013 Sep 30. Mol Microbiol. 2013. PMID: 24033548
-
[EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis].Seikagaku. 2010 Sep;82(9):825-31. Seikagaku. 2010. PMID: 20960918 Review. Japanese. No abstract available.
-
Mechanisms of ribosome recycling in bacteria and mitochondria: a structural perspective.RNA Biol. 2022;19(1):662-677. doi: 10.1080/15476286.2022.2067712. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35485608 Free PMC article. Review.
Cited by
-
Mitochondrial ribosomes in cancer.Semin Cancer Biol. 2017 Dec;47:67-81. doi: 10.1016/j.semcancer.2017.04.004. Epub 2017 Apr 23. Semin Cancer Biol. 2017. PMID: 28445780 Free PMC article. Review.
-
New compounds able to control hepatic cholesterol metabolism: Is it possible to avoid statin treatment in aged people?World J Hepatol. 2013 Dec 27;5(12):676-84. doi: 10.4254/wjh.v5.i12.676. World J Hepatol. 2013. PMID: 24432184 Free PMC article. Review.
References
-
- Coenen MJ, Antonicka H, Ugalde C, Sasarman F, Rossi R. Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med. 2004;351:2086. - PubMed
-
- Antonicka H, Sasarman F, Kennaway NG, Shoubridge EA. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum Mol Genet. 2006;15:1846. - PubMed
-
- Vladutiu G, Simmons Z, Isackson P, Tarnopolsky M, Peltier W. Genetic risk factors associated with lipid-lowering drug-induced myopathies. Muscle Nerve. 2006;32:162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

