A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy
- PMID: 22719832
- PMCID: PMC3374618
- DOI: 10.1371/journal.pone.0036466
A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy
Abstract
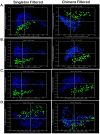
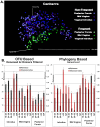
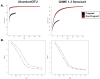
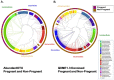
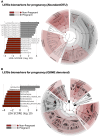
While current major national research efforts (i.e., the NIH Human Microbiome Project) will enable comprehensive metagenomic characterization of the adult human microbiota, how and when these diverse microbial communities take up residence in the host and during reproductive life are unexplored at a population level. Because microbial abundance and diversity might differ in pregnancy, we sought to generate comparative metagenomic signatures across gestational age strata. DNA was isolated from the vagina (introitus, posterior fornix, midvagina) and the V5V3 region of bacterial 16S rRNA genes were sequenced (454FLX Titanium platform). Sixty-eight samples from 24 healthy gravidae (18 to 40 confirmed weeks) were compared with 301 non-pregnant controls (60 subjects). Generated sequence data were quality filtered, taxonomically binned, normalized, and organized by phylogeny and into operational taxonomic units (OTU); principal coordinates analysis (PCoA) of the resultant beta diversity measures were used for visualization and analysis in association with sample clinical metadata. Altogether, 1.4 gigabytes of data containing >2.5 million reads (averaging 6,837 sequences/sample of 493 nt in length) were generated for computational analyses. Although gravidae were not excluded by virtue of a posterior fornix pH >4.5 at the time of screening, unique vaginal microbiome signature encompassing several specific OTUs and higher-level clades was nevertheless observed and confirmed using a combination of phylogenetic, non-phylogenetic, supervised, and unsupervised approaches. Both overall diversity and richness were reduced in pregnancy, with dominance of Lactobacillus species (L. iners crispatus, jensenii and johnsonii, and the orders Lactobacillales (and Lactobacillaceae family), Clostridiales, Bacteroidales, and Actinomycetales. This intergroup comparison using rigorous standardized sampling protocols and analytical methodologies provides robust initial evidence that the vaginal microbial 16S rRNA gene catalogue uniquely differs in pregnancy, with variance of taxa across vaginal subsite and gestational age.
Conflict of interest statement
Figures
References
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

