Different molecular signatures in magnetic resonance imaging-staged facioscapulohumeral muscular dystrophy muscles
- PMID: 22719944
- PMCID: PMC3374833
- DOI: 10.1371/journal.pone.0038779
Different molecular signatures in magnetic resonance imaging-staged facioscapulohumeral muscular dystrophy muscles
Abstract
Background: Facioscapulohumeral muscular dystrophy (FSHD) is one of the most common muscular dystrophies and is characterized by a non-conventional genetic mechanism activated by pathogenic D4Z4 repeat contractions. By muscle Magnetic Resonance Imaging (MRI) we observed that T2-short tau inversion recovery (T2-STIR) sequences identify two different conditions in which each muscle can be found before the irreversible dystrophic alteration, marked as T1-weighted sequence hyperintensity, takes place. We studied these conditions in order to obtain further information on the molecular mechanisms involved in the selective wasting of single muscles or muscle groups in this disease.
Methods: Histopathology, gene expression profiling and real time PCR were performed on biopsies from FSHD muscles with different MRI pattern (T1-weighted normal/T2-STIR normal and T1-weighted normal/T2-STIR hyperintense). Data were compared with those from inflammatory myopathies, dysferlinopathies and normal controls. In order to validate obtained results, two additional FSHD samples with different MRI pattern were analyzed.
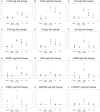
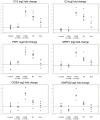
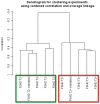
Results: Myopathic and inflammatory changes characterized T2-STIR hyperintense FSHD muscles, at variance with T2-STIR normal muscles. These two states could be easily distinguished from each other by their transcriptional profile. The comparison between T2-STIR hyperintense FSHD muscles and inflammatory myopathy muscles showed peculiar changes, although many alterations were shared among these conditions.
Conclusions: At the single muscle level, different stages of the disease correspond to the two MRI patterns. T2-STIR hyperintense FSHD muscles are more similar to inflammatory myopathies than to T2-STIR normal FSHD muscles or other muscular dystrophies, and share with them upregulation of genes involved in innate and adaptive immunity. Our data suggest that selective inflammation, together with perturbation in biological processes such as neoangiogenesis, lipid metabolism and adipokine production, may contribute to the sequential bursts of muscle degeneration that involve individual muscles in an asynchronous manner in this disease.
Conflict of interest statement
Figures


References
-
- Padberg GW, van Engelen BG. Facioscapulohumeral muscular dystrophy. Curr Opin Neurol. 2009;22:539–542. - PubMed
-
- Ricci E, Galluzzi G, Deidda G, Cacurri S, Colantoni L, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol. 1999;45:751–757. - PubMed
-
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. - PubMed
-
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

