How many protein-protein interactions types exist in nature?
- PMID: 22719985
- PMCID: PMC3374795
- DOI: 10.1371/journal.pone.0038913
How many protein-protein interactions types exist in nature?
Abstract
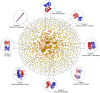
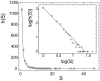
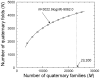
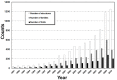
"Protein quaternary structure universe" refers to the ensemble of all protein-protein complexes across all organisms in nature. The number of quaternary folds thus corresponds to the number of ways proteins physically interact with other proteins. This study focuses on answering two basic questions: Whether the number of protein-protein interactions is limited and, if yes, how many different quaternary folds exist in nature. By all-to-all sequence and structure comparisons, we grouped the protein complexes in the protein data bank (PDB) into 3,629 families and 1,761 folds. A statistical model was introduced to obtain the quantitative relation between the numbers of quaternary families and quaternary folds in nature. The total number of possible protein-protein interactions was estimated around 4,000, which indicates that the current protein repository contains only 42% of quaternary folds in nature and a full coverage needs approximately a quarter century of experimental effort. The results have important implications to the protein complex structural modeling and the structure genomics of protein-protein interactions.
Conflict of interest statement
Figures
References
-
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. - PubMed
-
- Zhang C, DeLisi C. Estimating the number of protein folds. J Mol Biol. 1998;284:1301–1305. - PubMed
-
- Govindarajan S, Recabarren R, Goldstein RA. Estimating the total number of protein folds. Proteins. 1999;35:408–414. - PubMed
-
- Liu X, Fan K, Wang W. The number of protein folds and their distribution over families in nature. Proteins. 2004;54:491–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

