Temporal network based analysis of cell specific vein graft transcriptome defines key pathways and hub genes in implantation injury
- PMID: 22720046
- PMCID: PMC3376111
- DOI: 10.1371/journal.pone.0039123
Temporal network based analysis of cell specific vein graft transcriptome defines key pathways and hub genes in implantation injury
Abstract
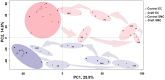
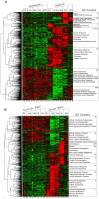
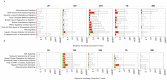
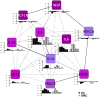
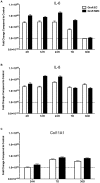
Vein graft failure occurs between 1 and 6 months after implantation due to obstructive intimal hyperplasia, related in part to implantation injury. The cell-specific and temporal response of the transcriptome to vein graft implantation injury was determined by transcriptional profiling of laser capture microdissected endothelial cells (EC) and medial smooth muscle cells (SMC) from canine vein grafts, 2 hours (H) to 30 days (D) following surgery. Our results demonstrate a robust genomic response beginning at 2 H, peaking at 12-24 H, declining by 7 D, and resolving by 30 D. Gene ontology and pathway analyses of differentially expressed genes indicated that implantation injury affects inflammatory and immune responses, apoptosis, mitosis, and extracellular matrix reorganization in both cell types. Through backpropagation an integrated network was built, starting with genes differentially expressed at 30 D, followed by adding upstream interactive genes from each prior time-point. This identified significant enrichment of IL-6, IL-8, NF-κB, dendritic cell maturation, glucocorticoid receptor, and Triggering Receptor Expressed on Myeloid Cells (TREM-1) signaling, as well as PPARα activation pathways in graft EC and SMC. Interactive network-based analyses identified IL-6, IL-8, IL-1α, and Insulin Receptor (INSR) as focus hub genes within these pathways. Real-time PCR was used for the validation of two of these genes: IL-6 and IL-8, in addition to Collagen 11A1 (COL11A1), a cornerstone of the backpropagation. In conclusion, these results establish causality relationships clarifying the pathogenesis of vein graft implantation injury, and identifying novel targets for its prevention.
Conflict of interest statement
Figures


References
-
- Hall M DC, Williams S, Golosinskiy A, Schwartzman A. National Hospital Discharge Survey: 2007 Summary. US Department of Health and Human Services. 2010. - PubMed
-
- Nowygrod R, Egorova N, Greco G, Anderson P, Gelijns A, et al. Trends, complications, and mortality in peripheral vascular surgery. J Vasc Surg. 2006;43:205–216. - PubMed
-
- DHHS. Statistics on Hospital-Based Care in the United States, 2007. The Healthcare Cost and Utilization Project: Department of Health and Human Services, USA. 2010.
-
- Conte M. Molecular engineering of vein bypass grafts. J Vasc Surg. 2007;45:A74–81. - PubMed
-
- Belkin M, Whittemore AD. Rutherford RB, editor. Infrainguinal bypass. 2000. pp. 98–1018. editor. Vascular surgery 5th ed. Philadelphia: WB Saunders Co.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

