Lymphoblastoid cell line with B1 cell characteristics established from a chronic lymphocytic leukemia clone by in vitro EBV infection
- PMID: 22720208
- PMCID: PMC3376971
- DOI: 10.4161/onci.1.1.18400
Lymphoblastoid cell line with B1 cell characteristics established from a chronic lymphocytic leukemia clone by in vitro EBV infection
Abstract
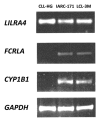
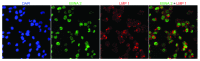
Chronic lymphocytic leukemia (CLL) cells express the receptor for Epstein-Barr virus (EBV) and can be infected in vitro. Infected cells do not express the growth-promoting set of EBV-encoded genes and therefore they do not yield LCLs, in most experiments. With exceptional clones, lines were obtained however. We describe a new line, HG3, established by in vitro EBV-infection from an IGHV1-2 unmutated CLL patient clone. All cells expressed EBNA-2 and LMP-1, the EBV-encoded genes pivotal for transformation. The karyotype, FISH cytogenetics and SNP-array profile of the line and the patient's ex vivo clone showed biallelic 13q14 deletions with genomic loss of DLEU7, miR15a/miR16-1, the two micro-RNAs that are deleted in 50% of CLL cases. Further features of CLL cells were: expression of CD5/CD20/CD27/CD43 and release of IgM natural antibodies reacting with oxLDL-like epitopes on apoptotic cells (cf. stereotyped subset-1). Comparison with two LCLs established from normal B cells showed 32 genes expressed at higher levels (> 2-fold). Among these were LHX2 and LILRA. These genes may play a role in the development of the disease. LHX2 expression was shown in self-renewing multipotent hematopoietic stem cells, and LILRA4 codes for a receptor for bone marrow stromal cell antigen-2 that contributes to B cell development. Twenty-four genes were expressed at lower levels, among these PARD3 that is essential for asymmetric cell division. These genes may contribute to establish precursors of CLL clones by regulation of cellular phenotype in the hematopoietic compartment. Expression of CD5/CD20/CD27/CD43 and spontaneous production of natural antibodies may identify the CLL cell as a self-renewing B1 lymphocyte.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
