Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates
- PMID: 22720735
- PMCID: PMC3438084
- DOI: 10.1186/1471-2180-12-116
Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates
Abstract
Background: Plant root exudates have been shown to play an important role in mediating interactions between plant growth-promoting rhizobacteria (PGPR) and their host plants. Most investigations were performed on Gram-negative rhizobacteria, while much less is known about Gram-positive rhizobacteria. To elucidate early responses of PGPR to root exudates, we investigated changes in the transcriptome of a Gram-positive PGPR to plant root exudates.
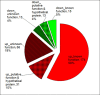
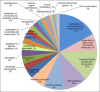
Results: Bacillus amyloliquefaciens FZB42 is a well-studied Gram-positive PGPR. To obtain a comprehensive overview of FZB42 gene expression in response to maize root exudates, microarray experiments were performed. A total of 302 genes representing 8.2% of the FZB42 transcriptome showed significantly altered expression levels in the presence of root exudates. The majority of the genes (261) was up-regulated after incubation of FZB42 with root exudates, whereas only 41 genes were down-regulated. Several groups of the genes which were strongly induced by the root exudates are involved in metabolic pathways relating to nutrient utilization, bacterial chemotaxis and motility, and non-ribosomal synthesis of antimicrobial peptides and polyketides.
Conclusions: Here we present a transcriptome analysis of the root-colonizing bacterium Bacillus amyloliquefaciens FZB42 in response to maize root exudates. The 302 genes identified as being differentially transcribed are proposed to be involved in interactions of Gram-positive bacteria with plants.
Figures




Similar articles
-
Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates.BMC Genomics. 2015 Sep 7;16(1):685. doi: 10.1186/s12864-015-1825-5. BMC Genomics. 2015. PMID: 26346121 Free PMC article.
-
Gene expression regulation in the plant growth promoting Bacillus atrophaeus UCMB-5137 stimulated by maize root exudates.Gene. 2016 Sep 15;590(1):18-28. doi: 10.1016/j.gene.2016.05.045. Epub 2016 Jun 1. Gene. 2016. PMID: 27259668
-
Influence of root exudates on the extracellular proteome of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42.Microbiology (Reading). 2015 Jan;161(Pt 1):131-147. doi: 10.1099/mic.0.083576-0. Epub 2014 Oct 29. Microbiology (Reading). 2015. PMID: 25355936
-
Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review.Front Microbiol. 2015 Jul 28;6:780. doi: 10.3389/fmicb.2015.00780. eCollection 2015. Front Microbiol. 2015. PMID: 26284057 Free PMC article. Review.
-
Bacillus velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol.Front Microbiol. 2018 Oct 16;9:2491. doi: 10.3389/fmicb.2018.02491. eCollection 2018. Front Microbiol. 2018. PMID: 30386322 Free PMC article. Review.
Cited by
-
Linking plant nutritional status to plant-microbe interactions.PLoS One. 2013 Jul 16;8(7):e68555. doi: 10.1371/journal.pone.0068555. Print 2013. PLoS One. 2013. PMID: 23874669 Free PMC article.
-
The Bacillus cereus Strain EC9 Primes the Plant Immune System for Superior Biocontrol of Fusarium oxysporum.Plants (Basel). 2022 Mar 2;11(5):687. doi: 10.3390/plants11050687. Plants (Basel). 2022. PMID: 35270157 Free PMC article.
-
Microbial expression profiles in the rhizosphere of willows depend on soil contamination.ISME J. 2014 Feb;8(2):344-58. doi: 10.1038/ismej.2013.163. Epub 2013 Sep 26. ISME J. 2014. PMID: 24067257 Free PMC article.
-
Genetic, Epigenetic and Phenotypic Diversity of Four Bacillus velezensis Strains Used for Plant Protection or as Probiotics.Front Microbiol. 2019 Nov 15;10:2610. doi: 10.3389/fmicb.2019.02610. eCollection 2019. Front Microbiol. 2019. PMID: 31803155 Free PMC article.
-
Transcriptomic profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in response to Mimosa pudica root exudates illuminates the molecular basis of their nodulation competitiveness and symbiotic evolutionary history.BMC Genomics. 2018 Jan 30;19(1):105. doi: 10.1186/s12864-018-4487-2. BMC Genomics. 2018. PMID: 29378510 Free PMC article.
References
-
- Kloepper JW, Schroth MN. Proc of the 4th Internat Conf on Plant Pathogenic Bacter. INRA, Angers, France; 1978. Plant growth-promoting rhizobacteria on radishes.
-
- Domenech J, Reddy MS, Kloepper JW, Ramos B, Gutierrez-Manero J. Combined application of the biological product LS213 with Bacillus, Pseudomonas or Chryseobacterium for growth promotion and biological control of soil-borne diseases in pepper and tomato. BioControl. 2006;51(2):245–258. doi: 10.1007/s10526-005-2940-z. - DOI
-
- Dessaux Y, Ryan PR, Thomashow LS, Weller DM. Rhizosphere engineering and management for sustainable agriculture. Plant Soil. 2009;321(1–2):363–383.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

