Whole-genome analysis informs breast cancer response to aromatase inhibition
- PMID: 22722193
- PMCID: PMC3383766
- DOI: 10.1038/nature11143
Whole-genome analysis informs breast cancer response to aromatase inhibition
Abstract
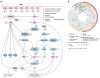
To correlate the variable clinical features of oestrogen-receptor-positive breast cancer with somatic alterations, we studied pretreatment tumour biopsies accrued from patients in two studies of neoadjuvant aromatase inhibitor therapy by massively parallel sequencing and analysis. Eighteen significantly mutated genes were identified, including five genes (RUNX1, CBFB, MYH9, MLL3 and SF3B1) previously linked to haematopoietic disorders. Mutant MAP3K1 was associated with luminal A status, low-grade histology and low proliferation rates, whereas mutant TP53 was associated with the opposite pattern. Moreover, mutant GATA3 correlated with suppression of proliferation upon aromatase inhibitor treatment. Pathway analysis demonstrated that mutations in MAP2K4, a MAP3K1 substrate, produced similar perturbations as MAP3K1 loss. Distinct phenotypes in oestrogen-receptor-positive breast cancer are associated with specific patterns of somatic mutations that map into cellular pathways linked to tumour biology, but most recurrent mutations are relatively infrequent. Prospective clinical trials based on these findings will require comprehensive genome sequencing.
Figures

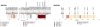


Comment in
-
Cancer genomics: indicators for drug response from sequencing.Nat Rev Genet. 2012 Jun 19;13(8):520. doi: 10.1038/nrg3285. Nat Rev Genet. 2012. PMID: 22710420 No abstract available.
-
Genomics: the breast cancer landscape.Nature. 2012 Jun 20;486(7403):328-9. doi: 10.1038/486328a. Nature. 2012. PMID: 22722187 No abstract available.
-
Genes, genes everywhere..Nat Rev Cancer. 2012 Jul 5;12(8):507. doi: 10.1038/nrc3323. Nat Rev Cancer. 2012. PMID: 22763664 No abstract available.
-
Who's driving anyway? Herculean efforts to identify the drivers of breast cancer.Breast Cancer Res. 2012 Oct 31;14(5):323. doi: 10.1186/bcr3325. Breast Cancer Res. 2012. PMID: 23113888 Free PMC article.
References
-
- Ellis MJ, et al. Randomized Phase II Neoadjuvant Comparison Between Letrozole, Anastrozole, and Exemestane for Postmenopausal Women With Estrogen Receptor-Rich Stage 2 to 3 Breast Cancer: Clinical and Biomarker Outcomes and Predictive Value of the Baseline PAM50-Based Intrinsic Subtype--ACOSOG Z1031. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.31.6950. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- P50 CA094056/CA/NCI NIH HHS/United States
- P50 CA068438/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 CA114722/CA/NCI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U24 CA196171/CA/NCI NIH HHS/United States
- U10 CA076001/CA/NCI NIH HHS/United States
- R01 CA095614/CA/NCI NIH HHS/United States
- 3P50 CA68438/CA/NCI NIH HHS/United States
- P50 CA94056/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

