Sequence analysis of mutations and translocations across breast cancer subtypes
- PMID: 22722202
- PMCID: PMC4148686
- DOI: 10.1038/nature11154
Sequence analysis of mutations and translocations across breast cancer subtypes
Abstract
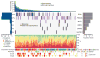
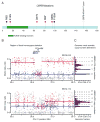
Breast carcinoma is the leading cause of cancer-related mortality in women worldwide, with an estimated 1.38 million new cases and 458,000 deaths in 2008 alone. This malignancy represents a heterogeneous group of tumours with characteristic molecular features, prognosis and responses to available therapy. Recurrent somatic alterations in breast cancer have been described, including mutations and copy number alterations, notably ERBB2 amplifications, the first successful therapy target defined by a genomic aberration. Previous DNA sequencing studies of breast cancer genomes have revealed additional candidate mutations and gene rearrangements. Here we report the whole-exome sequences of DNA from 103 human breast cancers of diverse subtypes from patients in Mexico and Vietnam compared to matched-normal DNA, together with whole-genome sequences of 22 breast cancer/normal pairs. Beyond confirming recurrent somatic mutations in PIK3CA, TP53, AKT1, GATA3 and MAP3K1, we discovered recurrent mutations in the CBFB transcription factor gene and deletions of its partner RUNX1. Furthermore, we have identified a recurrent MAGI3-AKT3 fusion enriched in triple-negative breast cancer lacking oestrogen and progesterone receptors and ERBB2 expression. The MAGI3-AKT3 fusion leads to constitutive activation of AKT kinase, which is abolished by treatment with an ATP-competitive AKT small-molecule inhibitor.
Figures

Comment in
-
Genomics: the breast cancer landscape.Nature. 2012 Jun 20;486(7403):328-9. doi: 10.1038/486328a. Nature. 2012. PMID: 22722187 No abstract available.
-
Genes, genes everywhere..Nat Rev Cancer. 2012 Jul 5;12(8):507. doi: 10.1038/nrc3323. Nat Rev Cancer. 2012. PMID: 22763664 No abstract available.
-
Who's driving anyway? Herculean efforts to identify the drivers of breast cancer.Breast Cancer Res. 2012 Oct 31;14(5):323. doi: 10.1186/bcr3325. Breast Cancer Res. 2012. PMID: 23113888 Free PMC article.
-
MAGI3-AKT3 fusion in breast cancer amended.Nature. 2015 Apr 16;520(7547):E11-2. doi: 10.1038/nature14265. Nature. 2015. PMID: 25877206 No abstract available.
-
Pugh et al. reply.Nature. 2015 Apr 16;520(7547):E12-4. doi: 10.1038/nature14266. Nature. 2015. PMID: 25877207 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

