Histone demethylase Kdm4b functions as a co-factor of C/EBPβ to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes
- PMID: 22722334
- PMCID: PMC3504706
- DOI: 10.1038/cdd.2012.75
Histone demethylase Kdm4b functions as a co-factor of C/EBPβ to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes
Abstract
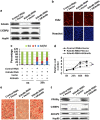
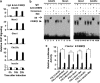
CCAAT/enhancer-binding protein (C/EBP) β is required for both mitotic clonal expansion (MCE) and terminal adipocyte differentiation of 3T3-L1 preadipocytes. Although the role of C/EBPβ in terminal adipocyte differentiation is well defined, its mechanism of action during MCE is not. In this report, histone demethylase Kdm4b, as well as cell cycle genes Cdc45l (cell division cycle 45 homolog), Mcm3 (mini-chromosome maintenance complex component 3), Gins1 (GINS complex subunit 1) and Cdc25c (cell division cycle 25 homolog c), were identified as potential C/EBPβ target genes during MCE by utilizing promoter-wide chromatin immunoprecipitation (ChIP)-on-chip analysis combined with gene expression microarrays. The expression of Kdm4b is induced during MCE and its induction is dependent on C/EBPβ. ChIP, Electrophoretic Mobility Shift Assay (EMSA) and luciferase assay confirmed that the promoter of Kdm4b is bound and activated by C/EBPβ. Knockdown of Kdm4b impaired MCE. Furthermore, Kdm4b interacted with C/EBPβ and was recruited to the promoters of C/EBPβ-regulated cell cycle genes, including Cdc45l, Mcm3, Gins1, and Cdc25c, demethylated H3K9me3 and activated their transcription. These findings suggest a novel feed forward mechanism involving a DNA binding transcription factor (C/EBPβ) and a chromatin regulator (Kdm4b) in the regulation of MCE by controlling cell cycle gene expression.
Figures






References
-
- Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. - PubMed
-
- Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268:22243–22246. - PubMed
-
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. - PubMed
-
- Qiu Z, Wei Y, Chen N, Jiang M, Wu J, Liao K. DNA synthesis and mitotic clonal expansion is not a required step for 3T3-L1 preadipocyte differentiation into adipocytes. J Biol Chem. 2001;276:11988–11995. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

