Ubiquitination and selective autophagy
- PMID: 22722335
- PMCID: PMC3524631
- DOI: 10.1038/cdd.2012.72
Ubiquitination and selective autophagy
Abstract
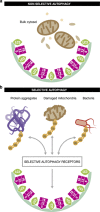
Ubiquitination has long been recognised as a key determinator of protein fate by tagging proteins for proteasomal degradation. Most recently, the ability of conjugated ubiquitin chains to confer selectivity to autophagy was demonstrated. Although autophagy was first believed to be a bulk, non-selective 'self-eating' degradative process, the molecular mechanisms of selectivity are now starting to emerge. With the discovery of autophagy receptors - which bind both ubiquitinated substrates and autophagy specific light chain 3 (LC3) modifier on the inner sheath of autophagosomes - a new pathway of selective autophagy is being unravelled. In this review, we focus on the special role of ubiquitin signals and selective autophagy receptors in sorting a variety of autophagic cargos.
Figures


References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. - PubMed
-
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

