The bonobo genome compared with the chimpanzee and human genomes
- PMID: 22722832
- PMCID: PMC3498939
- DOI: 10.1038/nature11128
The bonobo genome compared with the chimpanzee and human genomes
Abstract
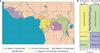
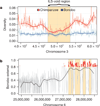
Two African apes are the closest living relatives of humans: the chimpanzee (Pan troglodytes) and the bonobo (Pan paniscus). Although they are similar in many respects, bonobos and chimpanzees differ strikingly in key social and sexual behaviours, and for some of these traits they show more similarity with humans than with each other. Here we report the sequencing and assembly of the bonobo genome to study its evolutionary relationship with the chimpanzee and human genomes. We find that more than three per cent of the human genome is more closely related to either the bonobo or the chimpanzee genome than these are to each other. These regions allow various aspects of the ancestry of the two ape species to be reconstructed. In addition, many of the regions that overlap genes may eventually help us understand the genetic basis of phenotypes that humans share with one of the two apes to the exclusion of the other.
Figures



References
-
- Boesch C, Hohmann G, Marchant L. Behavioural Diversity in Chimpanzees and Bonobos. Cambridge Univ. Press; 2002.
-
- de Waal F, Lanting F. Bonobo: the Forgotten Ape. Univ. California Press; 1997.
-
- Hare B, Wobber V, Wrangham R. The self-domestication hypothesis: evolution of bonobo psychology is due to selection against aggression. Anim. Behav. 2012;83:573–585.
-
- Kano T. The Last Ape: Pygmy Chimpanzee Behavior and Ecology. Stanford Univ. Press; 1992.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

