Proto-genes and de novo gene birth
- PMID: 22722833
- PMCID: PMC3401362
- DOI: 10.1038/nature11184
Proto-genes and de novo gene birth
Abstract
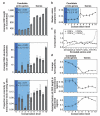
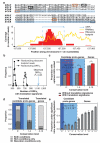
Novel protein-coding genes can arise either through re-organization of pre-existing genes or de novo. Processes involving re-organization of pre-existing genes, notably after gene duplication, have been extensively described. In contrast, de novo gene birth remains poorly understood, mainly because translation of sequences devoid of genes, or 'non-genic' sequences, is expected to produce insignificant polypeptides rather than proteins with specific biological functions. Here we formalize an evolutionary model according to which functional genes evolve de novo through transitory proto-genes generated by widespread translational activity in non-genic sequences. Testing this model at the genome scale in Saccharomyces cerevisiae, we detect translation of hundreds of short species-specific open reading frames (ORFs) located in non-genic sequences. These translation events seem to provide adaptive potential, as suggested by their differential regulation upon stress and by signatures of retention by natural selection. In line with our model, we establish that S. cerevisiae ORFs can be placed within an evolutionary continuum ranging from non-genic sequences to genes. We identify ~1,900 candidate proto-genes among S. cerevisiae ORFs and find that de novo gene birth from such a reservoir may be more prevalent than sporadic gene duplication. Our work illustrates that evolution exploits seemingly dispensable sequences to generate adaptive functional innovation.
Figures


Comment in
-
Evolution: the birth of new genes.Nat Rev Genet. 2012 Jul 3;13(8):521. doi: 10.1038/nrg3287. Nat Rev Genet. 2012. PMID: 22751427 No abstract available.
References
-
- Tautz D, Domazet-Loso T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011;12:692–702. - PubMed
-
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. - PubMed
-
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

