A map of nucleosome positions in yeast at base-pair resolution
- PMID: 22722846
- PMCID: PMC3786739
- DOI: 10.1038/nature11142
A map of nucleosome positions in yeast at base-pair resolution
Abstract
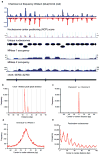
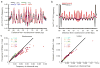
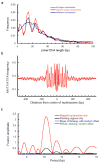
The exact positions of nucleosomes along genomic DNA can influence many aspects of chromosome function. However, existing methods for mapping nucleosomes do not provide the necessary single-base-pair accuracy to determine these positions. Here we develop and apply a new approach for direct mapping of nucleosome centres on the basis of chemical modification of engineered histones. The resulting map locates nucleosome positions genome-wide in unprecedented detail and accuracy. It shows new aspects of the in vivo nucleosome organization that are linked to transcription factor binding, RNA polymerase pausing and the higher-order structure of the chromatin fibre.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

