Heterogeneous pathways and timing of factor departure during translation initiation
- PMID: 22722848
- PMCID: PMC4465488
- DOI: 10.1038/nature11172
Heterogeneous pathways and timing of factor departure during translation initiation
Abstract
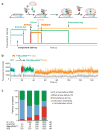
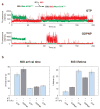
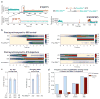
The initiation of translation establishes the reading frame for protein synthesis and is a key point of regulation. Initiation involves factor-driven assembly at a start codon of a messenger RNA of an elongation-competent 70S ribosomal particle (in bacteria) from separated 30S and 50S subunits and initiator transfer RNA. Here we establish in Escherichia coli, using direct single-molecule tracking, the timing of initiator tRNA, initiation factor 2 (IF2; encoded by infB) and 50S subunit joining during initiation. Our results show multiple pathways to initiation, with orders of arrival of tRNA and IF2 dependent on factor concentration and composition. IF2 accelerates 50S subunit joining and stabilizes the assembled 70S complex. Transition to elongation is gated by the departure of IF2 after GTP hydrolysis, allowing efficient arrival of elongator tRNAs to the second codon presented in the aminoacyl-tRNA binding site (A site). These experiments highlight the power of single-molecule approaches to delineate mechanisms in complex multicomponent systems.
Conflict of interest statement
Figures

Comment in
-
Ribosomes, start your engines.Nat Methods. 2012 Aug;9(8):780. doi: 10.1038/nmeth.2119. Nat Methods. 2012. PMID: 23019691 No abstract available.
References
-
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

