Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation
- PMID: 22722925
- PMCID: PMC3438927
- DOI: 10.1074/jbc.M112.366625
Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation
Abstract
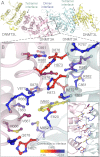
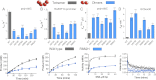
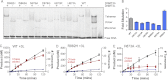
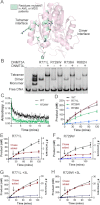
DNA methylation is a key regulator of gene expression and changes in DNA methylation occur early in tumorigenesis. Mutations in the de novo DNA methyltransferase gene, DNMT3A, frequently occur in adult acute myeloid leukemia patients with poor prognoses. Most of the mutations occur within the dimer or tetramer interface, including Arg-882. We have identified that the most prevalent mutation, R882H, and three additional mutants along the tetramer interface disrupt tetramerization. The processive methylation of multiple CpG sites is disrupted when tetramerization is eliminated. Our results provide a possible mechanism that accounts for how DNMT3A mutations may contribute to oncogenesis and its progression.
Figures



References
-
- Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 - PubMed
-
- Reik W., Dean W., Walter J. (2001) Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 - PubMed
-
- Robertson K. D. (2005) DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 - PubMed
-
- Okano M., Bell D. W., Haber D. A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 - PubMed
-
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. (2004) Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

