Characterization of Corynebacterium species in macaques
- PMID: 22723254
- PMCID: PMC3541768
- DOI: 10.1099/jmm.0.045377-0
Characterization of Corynebacterium species in macaques
Abstract
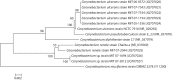
Bacteria of the genus Corynebacterium are important primary and opportunistic pathogens. Many are zoonotic agents. In this report, phenotypic (API Coryne analysis), genetic (rpoB and 16S rRNA gene sequencing), and physical methods (MS) were used to distinguish the closely related diphtheroid species Corynebacterium ulcerans and Corynebacterium pseudotuberculosis, and to definitively diagnose Corynebacterium renale from cephalic implants of rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques used in cognitive neuroscience research. Throat and cephalic implant cultures yielded 85 isolates from 43 macaques. Identification by API Coryne yielded C. ulcerans (n = 74), Corynebacterium pseudotuberculosis (n = 2), C. renale or most closely related to C. renale (n = 3), and commensals and opportunists (n = 6). The two isolates identified as C. pseudotuberculosis by API Coryne required genetic and MS analysis for accurate characterization as C. ulcerans. Of three isolates identified as C. renale by 16S rRNA gene sequencing, only one could be confirmed as such by API Coryne, rpoB gene sequencing and MS. This study emphasizes the importance of adjunct methods in identification of coryneforms and is the first isolation of C. renale from cephalic implants in macaques.
Figures

References
-
- Aubel D., Renaud F., Freney J. (1997). Genomic diversity of several Corynebacterium species identified by amplification of the 16S–23S rRNA gene spacer region. Int J Syst Bact 47, 767–772 10.1099/00207713-47-3-767 - DOI
-
- Bergin I. L., Chien C.-C., Marini R. P., Fox J. G. (2000). Isolation and characterization of Corynebacterium ulcerans from cephalic implants in macaques. Comp Med 50, 530–535 - PubMed
-
- De Zoysa A., Hawkey P. M., Engler K., George R., Mann G., Reilly W., Taylor D., Efstratiou A. (2005). Characterization of toxigenic Corynebacterium ulcerans strains isolated from humans and domestic cats in the United Kingdom. J Clin Microbiol 43, 4377–4381 10.1128/JCM.43.9.4377-4381.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

