Cytoplasmic RNA-binding proteins and the control of complex brain function
- PMID: 22723494
- PMCID: PMC3405866
- DOI: 10.1101/cshperspect.a012344
Cytoplasmic RNA-binding proteins and the control of complex brain function
Abstract
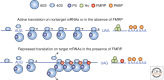
The formation and maintenance of neural circuits in the mammal central nervous system (CNS) require the coordinated expression of genes not just at the transcriptional level, but at the translational level as well. Recent evidence shows that regulated messenger RNA (mRNA) translation is necessary for certain forms of synaptic plasticity, the cellular basis of learning and memory. In addition, regulated translation helps guide axonal growth cones to their targets on other neurons or at the neuromuscular junction. Several neurologic syndromes have been correlated with and indeed may be caused by aberrant translation; one important example is the fragile X mental retardation syndrome. Although translation in the CNS is regulated by multiple mechanisms and factors, we focus this review on regulatory mRNA-binding proteins with particular emphasis on fragile X mental retardation protein (FMRP) and cytoplasmic polyadenylation element binding (CPEB) because they have been shown to be at the nexus of translational control and brain function in health and disease.
Figures

References
-
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H, et al. 1999. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci 96: 9885–9890 - PMC - PubMed
-
- Angenstein F, Evans AM, Ling SC, Settlage RE, Ficarro S, Carrero-Martinez FA, Shabanowitz J, Hunt DF, Greenough WT 2005. Proteomic characterization of messenger ribonucleoprotein complexes bound to nontranslated or translated poly(A) mRNAs in the rat cerebral cortex. J Biol Chem 280: 6496–6503 - PubMed
-
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ 2006. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci 32: 37–48 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
