Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis
- PMID: 22723521
- PMCID: PMC3392438
- DOI: 10.4049/jimmunol.1103031
Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis
Abstract
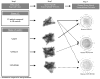
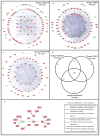
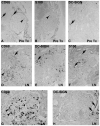
Lupus nephritis (LN) is a serious manifestation of systemic lupus erythematosus. Therapeutic studies in mouse LN models do not always predict outcomes of human therapeutic trials, raising concerns about the human relevance of these preclinical models. In this study, we used an unbiased transcriptional network approach to define, in molecular terms, similarities and differences among three lupus models and human LN. Genome-wide gene-expression networks were generated using natural language processing and automated promoter analysis and compared across species via suboptimal graph matching. The three murine models and human LN share both common and unique features. The 20 commonly shared network nodes reflect the key pathologic processes of immune cell infiltration/activation, endothelial cell activation/injury, and tissue remodeling/fibrosis, with macrophage/dendritic cell activation as a dominant cross-species shared transcriptional pathway. The unique nodes reflect differences in numbers and types of infiltrating cells and degree of remodeling among the three mouse strains. To define mononuclear phagocyte-derived pathways in human LN, gene sets activated in isolated NZB/W renal mononuclear cells were compared with human LN kidney profiles. A tissue compartment-specific macrophage-activation pattern was seen, with NF-κB1 and PPARγ as major regulatory nodes in the tubulointerstitial and glomerular networks, respectively. Our study defines which pathologic processes in murine models of LN recapitulate the key transcriptional processes active in human LN and suggests that there are functional differences between mononuclear phagocytes infiltrating different renal microenvironments.
Figures


References
-
- Doria A, Iaccarino L, Ghirardello A, Zampieri S, Arienti S, Sarzi-Puttini P, Atzeni F, Piccoli A, Todesco S. Long-term prognosis and causes of death in systemic lupus erythematosus. Am J Med. 2006;119:700–706. - PubMed
-
- Cook RJ, Gladman DD, Pericak D, Urowitz MB. Prediction of short term mortality in systemic lupus erythematosus with time dependent measures of disease activity. J Rheumatol. 2000;27:1892–1895. - PubMed
-
- Ward MM. Changes in the incidence of end-stage renal disease due to lupus nephritis, 1982–1995. Arch Intern Med. 2000;160:3136–3140. - PubMed
-
- Grande JP. Experimental models of lupus nephritis. Contrib Nephrol. 2011;169:183–197. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

