Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy
- PMID: 22723710
- PMCID: PMC3462658
- DOI: 10.1523/JNEUROSCI.0204-12.2012
Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy
Abstract
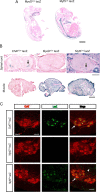
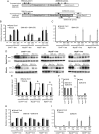
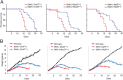
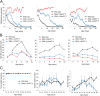
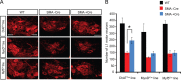
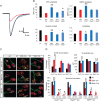
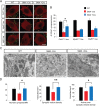
The inherited motor neuron disease spinal muscular atrophy (SMA) is caused by deficient expression of survival motor neuron (SMN) protein and results in severe muscle weakness. In SMA mice, synaptic dysfunction of both neuromuscular junctions (NMJs) and central sensorimotor synapses precedes motor neuron cell death. To address whether this synaptic dysfunction is due to SMN deficiency in motor neurons, muscle, or both, we generated three lines of conditional SMA mice with tissue-specific increases in SMN expression. All three lines of mice showed increased survival, weights, and improved motor behavior. While increased SMN expression in motor neurons prevented synaptic dysfunction at the NMJ and restored motor neuron somal synapses, increased SMN expression in muscle did not affect synaptic function although it did improve myofiber size. Together these data indicate that both peripheral and central synaptic integrity are dependent on motor neurons in SMA, but SMN may have variable roles in the maintenance of these different synapses. At the NMJ, it functions at the presynaptic terminal in a cell-autonomous fashion, but may be necessary for retrograde trophic signaling to presynaptic inputs onto motor neurons. Importantly, SMN also appears to function in muscle growth and/or maintenance independent of motor neurons. Our data suggest that SMN plays distinct roles in muscle, NMJs, and motor neuron somal synapses and that restored function of SMN at all three sites will be necessary for full recovery of muscle power.
Figures

Similar articles
-
Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene.J Neurosci. 2010 Sep 8;30(36):12005-19. doi: 10.1523/JNEUROSCI.2208-10.2010. J Neurosci. 2010. PMID: 20826664 Free PMC article.
-
Motor neuronal repletion of the NMJ organizer, Agrin, modulates the severity of the spinal muscular atrophy disease phenotype in model mice.Hum Mol Genet. 2017 Jul 1;26(13):2377-2385. doi: 10.1093/hmg/ddx124. Hum Mol Genet. 2017. PMID: 28379354 Free PMC article.
-
Improvement of neuromuscular synaptic phenotypes without enhanced survival and motor function in severe spinal muscular atrophy mice selectively rescued in motor neurons.PLoS One. 2013 Sep 23;8(9):e75866. doi: 10.1371/journal.pone.0075866. eCollection 2013. PLoS One. 2013. PMID: 24086650 Free PMC article.
-
At the "junction" of spinal muscular atrophy pathogenesis: the role of neuromuscular junction dysfunction in SMA disease progression.Curr Mol Med. 2013 Aug;13(7):1160-74. doi: 10.2174/15665240113139990044. Curr Mol Med. 2013. PMID: 23514457 Review.
-
Spinal muscular atrophy and the antiapoptotic role of survival of motor neuron (SMN) protein.Mol Neurobiol. 2013 Apr;47(2):821-32. doi: 10.1007/s12035-013-8399-5. Epub 2013 Jan 13. Mol Neurobiol. 2013. PMID: 23315303 Review.
Cited by
-
Neurotransmitter release in motor nerve terminals of a mouse model of mild spinal muscular atrophy.J Anat. 2014 Jan;224(1):74-84. doi: 10.1111/joa.12038. Epub 2013 Mar 13. J Anat. 2014. PMID: 23489475 Free PMC article.
-
Motor neuron biology and disease: A current perspective on infantile-onset spinal muscular atrophy.Future Neurol. 2018 Aug;13(3):161-172. doi: 10.2217/fnl-2018-0008. Epub 2018 Jul 6. Future Neurol. 2018. PMID: 31396020 Free PMC article.
-
Control of spinal motor neuron terminal differentiation through sustained Hoxc8 gene activity.Elife. 2022 Mar 22;11:e70766. doi: 10.7554/eLife.70766. Elife. 2022. PMID: 35315772 Free PMC article.
-
Motor defects in a Drosophila model for spinal muscular atrophy result from SMN depletion during early neurogenesis.PLoS Genet. 2022 Jul 25;18(7):e1010325. doi: 10.1371/journal.pgen.1010325. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35877682 Free PMC article.
-
SMN-targeted therapeutics for spinal muscular atrophy: are we SMArt enough yet?J Clin Invest. 2014 Feb;124(2):487-90. doi: 10.1172/JCI74142. Epub 2014 Jan 27. J Clin Invest. 2014. PMID: 24463455 Free PMC article.
References
-
- Biondi O, Grondard C, Lécolle S, Deforges S, Pariset C, Lopes P, Cifuentes-Diaz C, Li H, della Gaspera B, Chanoine C, Charbonnier F. Exercise-induced activation of NMDA receptor promotes motor unit development and survival in a type 2 spinal muscular atrophy model mouse. J Neurosci. 2008;28:953–962. - PMC - PubMed
-
- Braun S, Croizat B, Lagrange MC, Warter JM, Poindron P. Constitutive muscular abnormalities in culture in spinal muscular atrophy. Lancet. 1995;345:694–695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
