Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing
- PMID: 22723863
- PMCID: PMC3378559
- DOI: 10.1371/journal.pone.0038499
Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing
Abstract
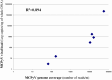
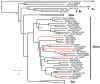
The human skin is a complex ecosystem that hosts a heterogeneous flora. Until recently, the diversity of the cutaneous microbiota was mainly investigated for bacteria through culture based assays subsequently confirmed by molecular techniques. There are now many evidences that viruses represent a significant part of the cutaneous flora as demonstrated by the asymptomatic carriage of beta and gamma-human papillomaviruses on the healthy skin. Furthermore, it has been recently suggested that some representatives of the Polyomavirus genus might share a similar feature. In the present study, the cutaneous virome of the surface of the normal-appearing skin from five healthy individuals and one patient with Merkel cell carcinoma was investigated through a high throughput metagenomic sequencing approach in an attempt to provide a thorough description of the cutaneous flora, with a particular focus on its viral component. The results emphasize the high diversity of the viral cutaneous flora with multiple polyomaviruses, papillomaviruses and circoviruses being detected on normal-appearing skin. Moreover, this approach resulted in the identification of new Papillomavirus and Circovirus genomes and confirmed a very low level of genetic diversity within human polyomavirus species. Although viruses are generally considered as pathogen agents, our findings support the existence of a complex viral flora present at the surface of healthy-appearing human skin in various individuals. The dynamics and anatomical variations of this skin virome and its variations according to pathological conditions remain to be further studied. The potential involvement of these viruses, alone or in combination, in skin proliferative disorders and oncogenesis is another crucial issue to be elucidated.
Conflict of interest statement
Figures

References
-
- Antonsson A, Erfurt C, Hazard K, Holmgren V, Simon M, et al. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol. 2003;84(7):1881–6. - PubMed
-
- Chen AC, McMillan NA, Antonsson A. Human papillomavirus type spectrum in normal skin of individuals with or without a history of frequent sun exposure. J Gen Virol. 2008;89(11):2891–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

