Deregulation of Rab and Rab effector genes in bladder cancer
- PMID: 22724020
- PMCID: PMC3378553
- DOI: 10.1371/journal.pone.0039469
Deregulation of Rab and Rab effector genes in bladder cancer
Abstract
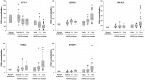
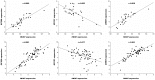
Growing evidence indicates that Rab GTPases, key regulators of intracellular transport in eukaryotic cells, play an important role in cancer. We analysed the deregulation at the transcriptional level of the genes encoding Rab proteins and Rab-interacting proteins in bladder cancer pathogenesis, distinguishing between the two main progression pathways so far identified in bladder cancer: the Ta pathway characterized by a high frequency of FGFR3 mutation and the carcinoma in situ pathway where no or infrequent FGFR3 mutations have been identified. A systematic literature search identified 61 genes encoding Rab proteins and 223 genes encoding Rab-interacting proteins. Transcriptomic data were obtained for normal urothelium samples and for two independent bladder cancer data sets corresponding to 152 and 75 tumors. Gene deregulation was analysed with the SAM (significant analysis of microarray) test or the binomial test. Overall, 30 genes were down-regulated, and 13 were up-regulated in the tumor samples. Five of these deregulated genes (LEPRE1, MICAL2, RAB23, STXBP1, SYTL1) were specifically deregulated in FGFR3-non-mutated muscle-invasive tumors. No gene encoding a Rab or Rab-interacting protein was found to be specifically deregulated in FGFR3-mutated tumors. Cluster analysis showed that the RAB27 gene cluster (comprising the genes encoding RAB27 and its interacting partners) was deregulated and that this deregulation was associated with both pathways of bladder cancer pathogenesis. Finally, we found that the expression of KIF20A and ZWINT was associated with that of proliferation markers and that the expression of MLPH, MYO5B, RAB11A, RAB11FIP1, RAB20 and SYTL2 was associated with that of urothelial cell differentiation markers. This systematic analysis of Rab and Rab effector gene deregulation in bladder cancer, taking relevant tumor subgroups into account, provides insight into the possible roles of Rab proteins and their effectors in bladder cancer pathogenesis. This approach is applicable to other group of genes and types of cancer.
Conflict of interest statement
Figures






References
-
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–25. - PubMed
-
- Lankat-Buttgereit B, Fehmann HC, Hering BJ, Bretzel RG, Göke B. Expression of the ras-related rab3a gene in human insulinomas and normal human pancreatic islets. Pancreas. 1994;9(4):434–8. - PubMed
-
- Amillet JM, Ferbus D, Real FX, Antony C, Muleris M, et al. Characterization of human Rab20 overexpressed in exocrine pancreatic carcinoma. Hum Pathol. 2006;37(3):256–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

