Impact of intestinal microbiota on intestinal luminal metabolome
- PMID: 22724057
- PMCID: PMC3380406
- DOI: 10.1038/srep00233
Impact of intestinal microbiota on intestinal luminal metabolome
Abstract
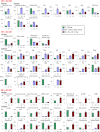
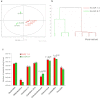
Low-molecular-weight metabolites produced by intestinal microbiota play a direct role in health and disease. In this study, we analyzed the colonic luminal metabolome using capillary electrophoresis mass spectrometry with time-of-flight (CE-TOFMS) -a novel technique for analyzing and differentially displaying metabolic profiles- in order to clarify the metabolite profiles in the intestinal lumen. CE-TOFMS identified 179 metabolites from the colonic luminal metabolome and 48 metabolites were present in significantly higher concentrations and/or incidence in the germ-free (GF) mice than in the Ex-GF mice (p < 0.05), 77 metabolites were present in significantly lower concentrations and/or incidence in the GF mice than in the Ex-GF mice (p < 0.05), and 56 metabolites showed no differences in the concentration or incidence between GF and Ex-GF mice. These indicate that intestinal microbiota highly influenced the colonic luminal metabolome and a comprehensive understanding of intestinal luminal metabolome is critical for clarifying host-intestinal bacterial interactions.
Conflict of interest statement
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry by the Bio-oriented Technology Research Advancement Institution (BRAIN), JAPAN. This work was funded by Kyodo Milk Industry Co. Ltd and Human Metabolome Technologies, Inc. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. M. Matsumoto and E. Sawaki are employees of Kyodo Milk Industry Co. Ltd. and had a role in study design, data analysis, preparation of the manuscript, and decision to publish the manuscript. T. Ooga is employee of Human Metabolome Technologies, Inc. and had a role in data analysis and decision to publish the manuscript. All of the other authors declare that they have no conflict of interest.
Figures


References
-
- Tannock G. W. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol 1 Suppl 1, S15–18 (2008). - PubMed
-
- Penders J., Stobberingh E. E., van den Brandt P. A. & Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy 62, 1223–1236 (2007). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

