RNA polymerase backtracking in gene regulation and genome instability
- PMID: 22726433
- PMCID: PMC3815583
- DOI: 10.1016/j.cell.2012.06.003
RNA polymerase backtracking in gene regulation and genome instability
Abstract
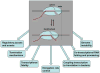
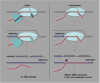
RNA polymerase is a ratchet machine that oscillates between productive and backtracked states at numerous DNA positions. Since its first description 15 years ago, backtracking--the reversible sliding of RNA polymerase along DNA and RNA--has been implicated in many critical processes in bacteria and eukaryotes, including the control of transcription elongation, pausing, termination, fidelity, and genome instability.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
-
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. - PubMed
-
- Alff-Steinberger C. A comparative study of mutations in Escherichia coli and Salmonella typhimurium shows that codon conservation is strongly correlated with codon usage. J Theor Biol. 2000;206:307–311. - PubMed
-
- Bailey M, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

