Genome-wide nucleosome specificity and directionality of chromatin remodelers
- PMID: 22726434
- PMCID: PMC3397793
- DOI: 10.1016/j.cell.2012.04.036
Genome-wide nucleosome specificity and directionality of chromatin remodelers
Abstract
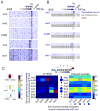
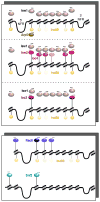
How chromatin remodelers cooperate to organize nucleosomes around the start and end of genes is not known. We determined the genome-wide binding of remodeler complexes SWI/SNF, RSC, ISW1a, ISW1b, ISW2, and INO80 to individual nucleosomes in Saccharomyces, and determined their functional contributions to nucleosome positioning through deletion analysis. We applied ultra-high-resolution ChIP-exo mapping to Isw2 to determine its subnucleosomal orientation and organization on a genomic scale. Remodelers interacted with selected nucleosome positions relative to the start and end of genes and produced net directionality in moving nucleosomes either away or toward nucleosome-free regions at the 5' and 3' ends of genes. Isw2 possessed a subnucleosomal organization in accord with biochemical and crystallographic-based models that place its linker binding region within promoters and abutted against Reb1-bound locations. Together, these findings reveal a coordinated position-specific approach taken by remodelers to organize genic nucleosomes into arrays.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Alen C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10:1441–1452. - PubMed
-
- Angermayr M, Oechsner U, Bandlow W. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J Biol Chem. 2003;278:17918–17926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

