Expressed pseudogenes in the transcriptional landscape of human cancers
- PMID: 22726445
- PMCID: PMC3597446
- DOI: 10.1016/j.cell.2012.04.041
Expressed pseudogenes in the transcriptional landscape of human cancers
Abstract
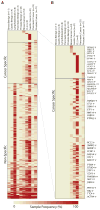
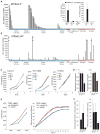
Pseudogene transcripts can provide a novel tier of gene regulation through generation of endogenous siRNAs or miRNA-binding sites. Characterization of pseudogene expression, however, has remained confined to anecdotal observations due to analytical challenges posed by the extremely close sequence similarity with their counterpart coding genes. Here, we describe a systematic analysis of pseudogene "transcription" from an RNA-Seq resource of 293 samples, representing 13 cancer and normal tissue types, and observe a surprisingly prevalent, genome-wide expression of pseudogenes that could be categorized as ubiquitously expressed or lineage and/or cancer specific. Further, we explore disease subtype specificity and functions of selected expressed pseudogenes. Taken together, we provide evidence that transcribed pseudogenes are a significant contributor to the transcriptional landscape of cells and are positioned to play significant roles in cellular differentiation and cancer progression, especially in light of the recently described ceRNA networks. Our work provides a transcriptome resource that enables high-throughput analyses of pseudogene expression.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Comment in
-
The great pretenders.Nat Rev Cancer. 2012 Jul 12;12(8):506. doi: 10.1038/nrc3326. Nat Rev Cancer. 2012. PMID: 22785353 No abstract available.
References
-
- Bier A, Oviedo-Landaverde I, Zhao J, Mamane Y, Kandouz M, Batist G. Connexin43 pseudogene in breast cancer cells offers a novel therapeutic target. Mol Cancer Ther. 2009;8:786–793. - PubMed
-
- Dormeyer W, van Hoof D, Braam SR, Heck AJ, Mummery CL, Krijgsveld J. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J Proteome Res. 2008;7:2936–2951. - PubMed
-
- Gang L, Janecka JE, Murphy WJ. Accelerated evolution of CES7, a gene encoding a novel major urinary protein in the Cat family. Mol Biol Evol. 2011;28:911–920. - PubMed
-
- Han H, Nutiu R, Moffat J, Blencowe BJ. SnapShot: High-throughput sequencing applications. Cell. 2011a;146:1044. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

