The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera
- PMID: 22726496
- PMCID: PMC3463433
- DOI: 10.1186/1471-2164-13-276
The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera
Abstract
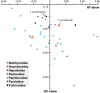
Background: Lepidoptera encompasses more than 160,000 described species that have been classified into 45-48 superfamilies. The previously determined Lepidoptera mitochondrial genomes (mitogenomes) are limited to six superfamilies of the lineage Ditrysia. Compared with the ancestral insect gene order, these mitogenomes all contain a tRNA rearrangement. To gain new insights into Lepidoptera mitogenome evolution, we sequenced the mitogenomes of two ghost moths that belong to the non-ditrysian lineage Hepialoidea and conducted a comparative mitogenomic analysis across Lepidoptera.
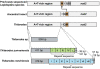
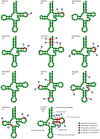
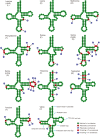
Results: The mitogenomes of Thitarodes renzhiensis and T. yunnanensis are 16,173 bp and 15,816 bp long with an A + T content of 81.28 % and 82.34 %, respectively. Both mitogenomes include 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and the A + T-rich region. Different tandem repeats in the A + T-rich region mainly account for the size difference between the two mitogenomes. All the protein-coding genes start with typical mitochondrial initiation codons, except for cox1 (CGA) and nad1 (TTG) in both mitogenomes. The anticodon of trnS(AGN) in T. renzhiensis and T. yunnanensis is UCU instead of the mostly used GCU in other sequenced Lepidoptera mitogenomes. The 1,584-bp sequence from rrnS to nad2 was also determined for an unspecified ghost moth (Thitarodes sp.), which has no repetitive sequence in the A + T-rich region. All three Thitarodes species possess the ancestral gene order with trnI-trnQ-trnM located between the A + T-rich region and nad2, which is different from the gene order trnM-trnI-trnQ in all previously sequenced Lepidoptera species. The formerly identified conserved elements of Lepidoptera mitogenomes (i.e. the motif 'ATAGA' and poly-T stretch in the A + T-rich region and the long intergenic spacer upstream of nad2) are absent in the Thitarodes mitogenomes.
Conclusion: The mitogenomes of T. renzhiensis and T. yunnanensis exhibit unusual features compared with the previously determined Lepidoptera mitogenomes. Their ancestral gene order indicates that the tRNA rearrangement event(s) likely occurred after Hepialoidea diverged from other lepidopteran lineages. Characterization of the two ghost moth mitogenomes has enriched our knowledge of Lepidoptera mitogenomes and contributed to our understanding of the mechanisms underlying mitogenome evolution, especially gene rearrangements.
Figures
References
-
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

