Celiac disease T-cell epitopes from gamma-gliadins: immunoreactivity depends on the genome of origin, transcript frequency, and flanking protein variation
- PMID: 22726570
- PMCID: PMC3469346
- DOI: 10.1186/1471-2164-13-277
Celiac disease T-cell epitopes from gamma-gliadins: immunoreactivity depends on the genome of origin, transcript frequency, and flanking protein variation
Abstract
Background: Celiac disease (CD) is caused by an uncontrolled immune response to gluten, a heterogeneous mixture of wheat storage proteins. The CD-toxicity of these proteins and their derived peptides is depending on the presence of specific T-cell epitopes (9-mer peptides; CD epitopes) that mediate the stimulation of HLA-DQ2/8 restricted T-cells. Next to the thoroughly characterized major T-cell epitopes derived from the α-gliadin fraction of gluten, γ-gliadin peptides are also known to stimulate T-cells of celiac disease patients. To pinpoint CD-toxic γ-gliadins in hexaploid bread wheat, we examined the variation of T-cell epitopes involved in CD in γ-gliadin transcripts of developing bread wheat grains.
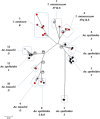
Results: A detailed analysis of the genetic variation present in γ-gliadin transcripts of bread wheat (T. aestivum, allo-hexaploid, carrying the A, B and D genome), together with genomic γ-gliadin sequences from ancestrally related diploid wheat species, enabled the assignment of sequence variants to one of the three genomic γ-gliadin loci, Gli-A1, Gli-B1 or Gli-D1. Almost half of the γ-gliadin transcripts of bread wheat (49%) was assigned to locus Gli-D1. Transcripts from each locus differed in CD epitope content and composition. The Gli-D1 transcripts contained the highest frequency of canonical CD epitope cores (on average 10.1 per transcript) followed by the Gli-A1 transcripts (8.6) and the Gli-B1 transcripts (5.4). The natural variants of the major CD epitope from γ-gliadins, DQ2-γ-I, showed variation in their capacity to induce in vitro proliferation of a DQ2-γ-I specific and HLA-DQ2 restricted T-cell clone.
Conclusions: Evaluating the CD epitopes derived from γ-gliadins in their natural context of flanking protein variation, genome specificity and transcript frequency is a significant step towards accurate quantification of the CD toxicity of bread wheat. This approach can be used to predict relative levels of CD toxicity of individual wheat cultivars directly from their transcripts (cDNAs).
Figures

References
-
- Dicke WK, Weijers NA, van der Kramer JH. Coeliac disease 2: The presence in wheat of a deleterious effect in cases of coeliac disease. Acta Paediatr. 1953;42:34. - PubMed
-
- Godkin A, Friede T, Davenport M, Stevanovic S, Willis A, Jewell D, Hill A, Rammensee HG. Use of eluted peptide sequence data to identify the binding characteristics of peptides to the insulin-dependent diabetes susceptibility allele HLA-DQ8 (DQ 3.2) Int Immunol. 1997;9:905–911. doi: 10.1093/intimm/9.6.905. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

