Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability
- PMID: 22726846
- PMCID: PMC3397275
- DOI: 10.1016/j.ajhg.2012.05.003
Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability
Abstract
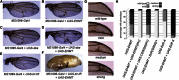
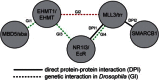
Intellectual disability (ID) disorders are genetically and phenotypically highly heterogeneous and present a major challenge in clinical genetics and medicine. Although many genes involved in ID have been identified, the etiology is unknown in most affected individuals. Moreover, the function of most genes associated with ID remains poorly characterized. Evidence is accumulating that the control of gene transcription through epigenetic modification of chromatin structure in neurons has an important role in cognitive processes and in the etiology of ID. However, our understanding of the key molecular players and mechanisms in this process is highly fragmentary. Here, we identify a chromatin-modification module that underlies a recognizable form of ID, the Kleefstra syndrome phenotypic spectrum (KSS). In a cohort of KSS individuals without mutations in EHMT1 (the only gene known to be disrupted in KSS until now), we identified de novo mutations in four genes, MBD5, MLL3, SMARCB1, and NR1I3, all of which encode epigenetic regulators. Using Drosophila, we demonstrate that MBD5, MLL3, and NR1I3 cooperate with EHMT1, whereas SMARCB1 is known to directly interact with MLL3. We propose a highly conserved epigenetic network that underlies cognition in health and disease. This network should allow the design of strategies to treat the growing group of ID pathologies that are caused by epigenetic defects.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. - PubMed
-
- van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu. Rev. Genet. 2011;45:81–104. - PubMed
-
- van Bokhoven H., Kramer J.M. Disruption of the epigenetic code: An emerging mechanism in mental retardation. Neurobiol. Dis. 2010;39:3–12. - PubMed
-
- Stewart D.R., Kleefstra T. The chromosome 9q subtelomere deletion syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2007;145C:383–392. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

