Immunoproteasomes: structure, function, and antigen presentation
- PMID: 22727420
- PMCID: PMC4405001
- DOI: 10.1016/B978-0-12-397863-9.00003-1
Immunoproteasomes: structure, function, and antigen presentation
Abstract
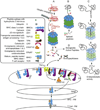
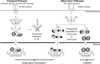
Immunoproteasomes contain replacements for the three catalytic subunits of standard proteasomes. In most cells, oxidative stress and proinflammatory cytokines are stimuli that lead to elevated production of immunoproteasomes. Immune system cells, especially antigen-presenting cells, express a higher basal level of immunoproteasomes. A well-described function of immunoproteasomes is to generate peptides with a hydrophobic C terminus that can be processed to fit in the groove of MHC class I molecules. This display of peptides on the cell surface allows surveillance by CD8 T cells of the adaptive immune system for pathogen-infected cells. Functions of immunoproteasomes, other than generating peptides for antigen presentation, are emerging from studies in immunoproteasome-deficient mice, and are complemented by recently described diseases linked to mutations or single-nucleotide polymorphisms in immunoproteasome subunits. Thus, this growing body of literature suggests a more pleiotropic role in cell function for the immunoproteasome.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


References
-
- Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. - PubMed
-
- Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355–357. - PubMed
-
- Glynne R, Powis SH, Beck S, Kelly A, Kerr LA, Trowsdale J. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature. 1991;353:357–360. - PubMed
-
- Kelly A, Powis SH, Glynne R, Radley E, Beck S, Trowsdale J. Second proteasome-related gene in the human MHC class II region. Nature. 1991;353:667–668. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

