Application of paramagnetic NMR-validated molecular dynamics simulation to the analysis of a conformational ensemble of a branched oligosaccharide
- PMID: 22728360
- PMCID: PMC6268797
- DOI: 10.3390/molecules17066658
Application of paramagnetic NMR-validated molecular dynamics simulation to the analysis of a conformational ensemble of a branched oligosaccharide
Abstract
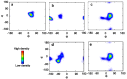
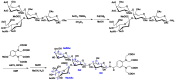
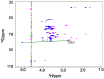
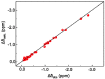
Oligosaccharides of biological importance often exhibit branched covalent structures and dynamic conformational multiplicities. Here we report the application of a method that we developed, which combined molecular dynamics (MD) simulations and lanthanide-assisted paramagnetic NMR spectroscopy, to evaluate the dynamic conformational ensemble of a branched oligosaccharide. A lanthanide-chelating tag was attached to the reducing end of the branched tetrasaccharide of GM2 ganglioside to observe pseudocontact shifts as the source of long distance information for validating the conformational ensemble derived from MD simulations. By inspecting the results, the conformational space of the GM2 tetrasaccharide was compared with that of its nonbranched derivative, the GM3 trisaccharide.
Figures





Similar articles
-
Paramagnetic NMR probes for characterization of the dynamic conformations and interactions of oligosaccharides.Glycoconj J. 2015 Oct;32(7):505-13. doi: 10.1007/s10719-015-9599-1. Epub 2015 Jun 7. Glycoconj J. 2015. PMID: 26050258 Review.
-
Lanthanide-assisted NMR evaluation of a dynamic ensemble of oligosaccharide conformations.Chem Commun (Camb). 2012 May 16;48(39):4752-4. doi: 10.1039/c2cc30353a. Epub 2012 Apr 4. Chem Commun (Camb). 2012. PMID: 22472911
-
Conformational Analysis of a High-Mannose-Type Oligosaccharide Displaying Glucosyl Determinant Recognised by Molecular Chaperones Using NMR-Validated Molecular Dynamics Simulation.Chembiochem. 2017 Feb 16;18(4):396-401. doi: 10.1002/cbic.201600595. Epub 2017 Jan 4. Chembiochem. 2017. PMID: 27995699
-
Exploration of conformational spaces of high-mannose-type oligosaccharides by an NMR-validated simulation.Angew Chem Int Ed Engl. 2014 Oct 6;53(41):10941-4. doi: 10.1002/anie.201406145. Epub 2014 Sep 4. Angew Chem Int Ed Engl. 2014. PMID: 25196214
-
Design and applications of lanthanide chelating tags for pseudocontact shift NMR spectroscopy with biomacromolecules.Prog Nucl Magn Reson Spectrosc. 2019 Oct-Dec;114-115:284-312. doi: 10.1016/j.pnmrs.2019.08.002. Epub 2019 Aug 28. Prog Nucl Magn Reson Spectrosc. 2019. PMID: 31779884 Review.
Cited by
-
Experimental and computational characterization of dynamic biomolecular interaction systems involving glycolipid glycans.Glycoconj J. 2022 Apr;39(2):219-228. doi: 10.1007/s10719-022-10056-w. Epub 2022 Mar 17. Glycoconj J. 2022. PMID: 35298725 Review.
-
Paramagnetic NMR probes for characterization of the dynamic conformations and interactions of oligosaccharides.Glycoconj J. 2015 Oct;32(7):505-13. doi: 10.1007/s10719-015-9599-1. Epub 2015 Jun 7. Glycoconj J. 2015. PMID: 26050258 Review.
-
Breaking the Limits in Analyzing Carbohydrate Recognition by NMR Spectroscopy: Resolving Branch-Selective Interaction of a Tetra-Antennary N-Glycan with Lectins.Angew Chem Int Ed Engl. 2017 Nov 20;56(47):14987-14991. doi: 10.1002/anie.201709130. Epub 2017 Oct 24. Angew Chem Int Ed Engl. 2017. PMID: 28991403 Free PMC article.
-
"Rules of Engagement" of Protein-Glycoconjugate Interactions: A Molecular View Achievable by using NMR Spectroscopy and Molecular Modeling.ChemistryOpen. 2016 Jun 7;5(4):274-96. doi: 10.1002/open.201600024. eCollection 2016 Aug. ChemistryOpen. 2016. PMID: 27547635 Free PMC article. Review.
-
Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.PLoS Pathog. 2015 Oct 16;11(10):e1005104. doi: 10.1371/journal.ppat.1005104. eCollection 2015 Oct. PLoS Pathog. 2015. PMID: 26474293 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

