Immunoglobulin class-switch DNA recombination: induction, targeting and beyond
- PMID: 22728528
- PMCID: PMC3545482
- DOI: 10.1038/nri3216
Immunoglobulin class-switch DNA recombination: induction, targeting and beyond
Abstract
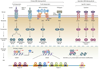
Class-switch DNA recombination (CSR) of the immunoglobulin heavy chain (IGH) locus is central to the maturation of the antibody response and crucially requires the cytidine deaminase AID. CSR involves changes in the chromatin state and the transcriptional activation of the IGH locus at the upstream and downstream switch (S) regions that are to undergo S-S DNA recombination. In addition, CSR involves the induction of AID expression and the targeting of CSR factors to S regions by 14-3-3 adaptors, and it is facilitated by the transcription machinery and by histone modifications. In this Review, we focus on recent advances regarding the induction and targeting of CSR and outline an integrated model of the assembly of macromolecular complexes that transduce crucial epigenetic information to enzymatic effectors of the CSR machinery.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

